Machine Learning-Accelerated Genetic Algorithms (MLaGA): Revolutionizing Nanoparticle Discovery for Biomedical Applications
This article explores the transformative integration of Machine Learning (ML) with Genetic Algorithms (GA) to accelerate the discovery and optimization of nanoparticles for drug delivery and biomedical applications.
Machine Learning-Accelerated Genetic Algorithms (MLaGA): Revolutionizing Nanoparticle Discovery for Biomedical Applications
Abstract
This article explores the transformative integration of Machine Learning (ML) with Genetic Algorithms (GA) to accelerate the discovery and optimization of nanoparticles for drug delivery and biomedical applications. Aimed at researchers, scientists, and drug development professionals, it provides a comprehensive examination of MLaGA foundations, from core principles to advanced methodologies. The content details practical applications in optimizing materials like PLGA and gold nanoparticles, addresses key challenges in troubleshooting and optimizing these computational workflows, and offers a critical validation against traditional methods. By synthesizing recent research and case studies, this article serves as a strategic guide for leveraging MLaGA to navigate vast combinatorial design spaces, significantly reduce development timelines, and usher in a new era of data-driven nanomedicine.
The Foundation of MLaGA: Core Principles and the Drive for Efficiency in Nanomedicine
In the pursuit of advanced therapeutics, the discovery and optimization of nanoparticles for drug delivery present a complex, multi-parameter challenge. Traditional experimental approaches are often slow, costly, and struggle to navigate the vast design space of material compositions, sizes, and surface properties. Machine Learning-accelerated Genetic Algorithms (MLaGAs) represent a powerful synergy of evolutionary computation and machine learning, engineered to overcome these limitations. This paradigm integrates the global search prowess of Genetic Algorithms (GAs) with the predictive modeling and pattern recognition capabilities of ML, creating a robust framework for intelligent design and optimization. Within nanoparticle discovery research, this hybrid approach is transforming the pace at which researchers can formulate stable, effective, and safe nanocarriers, de-risking the development pipeline and unlocking novel therapeutic possibilities [1] [2].
Theoretical Foundation
Core Components of the MLaGA Paradigm
A Machine Learning-accelerated Genetic Algorithm is an advanced optimization engine that uses machine learning models to enhance the efficiency and effectiveness of a traditional genetic algorithm. The core components of this hybrid paradigm are:
- Genetic Algorithm (GA): A population-based metaheuristic inspired by natural selection. It operates on a population of candidate solutions (e.g., different nanoparticle formulations), applying selection, crossover (recombination), and mutation operators to evolve increasingly fit solutions over generations [3]. GAs excel at global exploration of complex search spaces where gradient information is unavailable or the problem is a "black box."
- Machine Learning (ML) Model: A predictive algorithm, such as XGBoost or Support Vector Machines (SVM), that is integrated into the GA workflow [4] [3]. Its primary role is to act as a surrogate model, approximating the fitness function—which is often a costly and time-consuming wet-lab experiment. By rapidly predicting the performance of candidate formulations, the ML model drastically reduces the number of physical experiments required.
The Acceleration Mechanism
The "acceleration" in MLaGA is achieved by leveraging the ML model as a computationally cheap proxy for evaluation. Instead of synthesizing and testing every candidate nanoparticle in a lab, a large proportion of the GA population is evaluated using the ML model's prediction. This allows the algorithm to explore a much wider design space and converge to high-performing solutions in a fraction of the time. The ML model can be trained on initial experimental data and updated iteratively as new data is generated, progressively improving its predictive accuracy and guiding the GA more effectively [2] [3].
Performance and Quantitative Benchmarks
The superiority of hybrid approaches like MLaGA is demonstrated by their performance against traditional methods in both data generation and nanoparticle optimization tasks.
Table 1: Performance Comparison of Data Generation Techniques for Imbalanced Datasets
| Method | Accuracy | Precision | Recall | F1-Score | ROC-AUC |
|---|---|---|---|---|---|
| GA-based Synthesis [3] | Highest | Highest | Highest | Highest | Highest |
| XGBoost [4] | >96% | High | High | High | - |
| SMOTE [3] | <90% | Moderate | Moderate | Moderate | Lower |
| ADASYN [3] | <90% | Moderate | Moderate | Moderate | Lower |
| SVM / k-NN [4] | <90% | Lower | Lower | Lower | - |
Table 2: Efficacy of an AI-Guided Platform (TuNa-AI) in Nanoparticle Optimization
| Metric | Standard Approaches | TuNa-AI Platform [2] | Improvement |
|---|---|---|---|
| Successful Nanoparticle Formation | Baseline | +42.9% | 42.9% increase |
| Excipient Usage Reduction | Baseline | Reduced a carcinogenic excipient by 75% | Safer formulation |
| Therapeutic Efficacy | Baseline | Improved solubility and cell growth inhibition | Enhanced |
Application Notes: MLaGA for Nanoparticle Discovery
Primary Application Areas
In nanoparticle research, MLaGAs are primarily deployed for two critical tasks:
- Optimizing Formulation Recipes: The platform can simultaneously optimize both the identity of ingredients (e.g., polymers, lipids, drugs) and their quantitative ratios, a capability beyond many existing AI tools. This was validated in the development of a venetoclax-loaded nanoparticle for leukemia, which showed improved solubility and efficacy [2].
- Addressing Data Imbalance: In discovery screens, high-performing formulations are often rare, creating an imbalanced dataset. GAs can be used to generate high-quality synthetic data representing the minority class (high-performing nanoparticles), which then trains a more robust ML model for subsequent screening rounds [3].
Experimental Protocol: MLaGA-Driven Nanoparticle Optimization
The following workflow, implemented using an automated robotic platform and AI, outlines the process for discovering and optimizing a therapeutic nanoparticle formulation [2].
Step-by-Step Protocol:
- Define Design Space: Identify the variables to be optimized, including the types of therapeutic molecules, excipients (e.g., lipids, polymers), and their allowable concentration ranges [2].
- Generate Initial Library & High-Throughput Synthesis: Use an automated liquid handling robot to systematically prepare a library of distinct formulations (e.g., 1275 distinct formulations) by combining ingredients across the defined design space [2].
- Performance Assays: Characterize each formulation for critical performance attributes. Key assays include:
- Encapsulation Efficiency: Measure the percentage of the active drug successfully incorporated into the nanoparticle.
- Stability & Solubility: Assess physical stability in solution and dissolution profile.
- In Vitro Efficacy: Test the formulation's ability to inhibit target cell growth (e.g., leukemia cells) [2].
- Curate Training Dataset: Compile the data from steps 2 and 3 into a structured dataset where the input variables are the formulation recipes and the output/target variables are the performance metrics.
- Train Surrogate ML Model: Train a machine learning model (e.g., a hybrid kernel machine like TuNa-AI or a tree-based model like XGBoost) on the curated dataset. This model will learn to predict formulation performance based on the recipe [2].
- Run Genetic Algorithm:
- Initialization: Create an initial population of nanoparticle formulations.
- Evaluation: Use the trained surrogate ML model to rapidly predict the fitness (e.g., encapsulation efficiency, stability) of each candidate, instead of running a wet-lab experiment.
- Selection, Crossover, Mutation: Apply GA operators to create a new generation of candidate formulations [3].
- Iteration: Repeat the evaluation and evolution cycle for multiple generations.
- Select Promising Candidates: After the GA converges, select the top-performing candidate formulations from the final population.
- Experimental Validation: Synthesize and physically test the top candidates in the lab using the assays from Step 3 to confirm the ML model's predictions.
- Update Dataset & Model: Incorporate the new experimental results into the training dataset and retrain the ML model to improve its accuracy for subsequent optimization cycles.
- Deliver Optimized Formulation: The candidate that meets all pre-defined efficacy and safety criteria proceeds to further development.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Materials for MLaGA-Driven Nanoparticle Discovery
| Category / Item | Function / Purpose | Example Application |
|---|---|---|
| Therapeutic Molecules | The active pharmaceutical ingredient (API) to be delivered. | Venetoclax (for leukemia) [2]. |
| Excipients (Lipids, Polymers) | Inactive substances that form the nanoparticle structure, control drug release, and enhance stability. | PLGA, chitosan, lipids; optimized for safety and biodistribution [2] [5]. |
| Automated Liquid Handler | A robotic system for high-throughput, precise, and reproducible preparation of nanoparticle formulation libraries. | Systematic creation of 1275+ distinct formulations for initial dataset generation [2]. |
| AI/ML Software Stack | Software and algorithms for building surrogate models and running genetic algorithms. | EvoJAX, PyGAD for GAs; XGBoost, SVM for classification and regression [1] [4] [3]. |
| Characterization Assays | Experiments to measure nanoparticle properties and performance. | Encapsulation efficiency, stability/solubility tests, in vitro cell-based efficacy assays [2]. |
Advanced Protocol: Synthetic Data Generation for Imbalanced Formulation Data
A significant challenge in applying ML to nanoparticle discovery is the "imbalanced dataset" problem, where successful formulations are rare. This protocol uses a GA to generate synthetic data representing high-performing nanoparticles, which can then be used to augment training data for a secondary ML model.
Step-by-Step Protocol:
- Initial Imbalanced Dataset: Begin with a dataset of formulated nanoparticles where the minority class represents the successful/high-performing formulations.
- Fit ML Model to Minority Class: Train a machine learning model, such as a Support Vector Machine (SVM) or Logistic Regression, solely on the feature vectors of the minority class. This model learns the underlying distribution and patterns of the successful formulations [3].
- Derive Fitness Function: Use the trained ML model to create the fitness function for the GA. A candidate solution's (a synthetic data point's) fitness can be its proximity to the decision boundary of the SVM or its probability as assigned by the logistic regression model. The goal is to maximize this fitness, generating data that is convincingly part of the minority class [3].
- Initialize GA Population: Create an initial population of random candidate solutions (synthetic data vectors).
- Evaluate Fitness: Calculate the fitness of each candidate using the ML-derived fitness function.
- Apply GA Operators: Use selection, crossover, and mutation to create a new generation of candidates. Elitist GA variants, which preserve the best-performing candidates from one generation to the next, are often particularly effective [3].
- Iterate and Check Convergence: Repeat steps 5 and 6 until the population converges or a performance plateau is reached.
- Output and Augment: The final GA population represents a set of high-quality synthetic data for the minority class. This data is then combined with the original imbalanced dataset to create a balanced dataset, which is used to train a more robust and accurate final predictive model for nanoparticle screening.
Machine Learning-accelerated Genetic Algorithms represent a paradigm shift in computational design and optimization for nanoparticle discovery. By fusing the global search power of evolutionary algorithms with the predictive precision of machine learning, MLaGAs create a highly efficient, iterative discovery loop. This approach directly addresses two of the most significant bottlenecks in the field: the astronomical size of the possible formulation space and the scarcity of high-performance training data. As a result, MLaGAs empower researchers to extract latent value from complex biological and chemical constraints, de-risking the development pipeline and accelerating the creation of next-generation nanotherapeutics. The continued development and application of this hybrid paradigm promise to significantly shorten the timeline from therapeutic concept to viable, optimized nanoparticle drug product.
The Scale of the Combinatorial Problem
The design of nanoparticles, particularly for advanced applications like drug delivery, is governed by a vast number of interdependent parameters. These include, but are not limited to, core composition, size, shape, surface chemistry, and functionalization with targeting ligands. The number of possible combinations arising from these parameters creates a search space so large that it becomes practically impossible to explore exhaustively using traditional experimental methods.
For example, in the development of lipid nanoparticles (LNPs)—essential carriers for genetic medicines—optimization is plagued by nearly infinite design variables. Performance relies on a complex series of tasks:
- Encapsulation of nucleic acids
- Stable particle formation
- Stable circulation in the bloodstream
- Favorable interaction and endosomal uptake in target cells
- Endosomal escape to the cytoplasm [6].
Each of these tasks is influenced by subtle, interdependent changes to parameters such as lipid structure, lipid composition, cargo-to-vehicle material ratio, particle fabrication process, and surface surfactants [6]. This multi-scale and multi-parameter complexity makes leveraging computational power essential for rational design and optimization [6].
To put this into perspective, consider the challenge of optimizing a simple binary nanoalloy, such as a PtxAu147-x nanoparticle. The number of possible atomic arrangements, or homotops, for each composition is given by the combinatorial formula: $$HN = \frac{{N!}}{{N{\mathrm{A}}!N_{\mathrm{B}}!}}$$ For all 146 compositions, this results in a total of 1.78 × 10^44 homotops [7]. The number of possibilities rises combinatorially toward the 1:1 composition, making a brute-force search for the most stable structure entirely infeasible [7].
The Inefficiency of Traditional Optimization Methods
Traditional nanoparticle discovery often relies on a trial-and-error approach in the laboratory. This method is time-consuming, costly, and challenging, as it depends on finding the optimum formulation under controlled experimental conditions with high equipment supplies and practical experience [8]. These "brute-force" methods are inefficient because they require a large number of experiments or calculations and are often limited by the researcher's intuition and prior knowledge.
The impracticality is evident when considering computational searches. A traditional Genetic Algorithm (GA) used to locate the most stable structures (the convex hull) for the 147-atom Pt-Au system required approximately 16,000 candidate minimizations [7]. While this is significantly lower than the total number of homotops, it is still prohibitively high when using computationally expensive energy calculators, such as Density Functional Theory (DFT), which provides accurate results but is resource-intensive [7].
The following table quantifies the inefficiency of a traditional GA for a specific nanoalloy search and contrasts it with a machine-learning accelerated approach.
Table 1: Comparison of Search Methods for Locating the Convex Hull of PtxAu147-x Nanoparticles
| Search Method | Number of Energy Calculations Required | Computational Cost | Key Characteristics |
|---|---|---|---|
| Brute-Force | 1.78 x 10^44 (Total homotops) [7] | Theoretically impossible | Evaluates all possible combinations |
| Traditional Genetic Algorithm (GA) | ~16,000 [7] | High, often prohibitive for accurate methods like DFT | Metaheuristic, inspired by Darwinian evolution |
| Machine Learning Accelerated GA (MLaGA) | ~280 - 1,200 [7] | 50-fold reduction compared to traditional GA [7] [9] | Uses a machine learning model as a surrogate fitness evaluator |
This table demonstrates that even advanced metaheuristic algorithms like GAs can be inefficient, requiring thousands of expensive evaluations. This inefficiency forms the core of the combinatorial challenge, making the discovery of optimal nanoparticle designs slow and resource-intensive.
Experimental Protocol: Traditional "Brute-Force" Screening for Nanoparticle Properties
This protocol outlines a standardized, iterative process for experimentally determining the optimal formulation parameters for a polymer-based nanoparticle, such as Poly(lactic-co-glycolic acid) (PLGA), to achieve a target size and drug encapsulation efficiency.
Materials and Equipment
Table 2: Key Research Reagents and Equipment for Traditional Nanoparticle Screening
| Item Name | Function/Description |
|---|---|
| PLGA (50:50) | A biodegradable and biocompatible copolymer used as the primary matrix material for nanoparticle formation [8]. |
| Solvent (e.g., Acetone, DCM) | An organic solvent used to dissolve the polymer. |
| Aqueous Surfactant Solution (e.g., PVA) | A stabilizer that prevents nanoparticle aggregation during formation. |
| Antiviral Drug Candidate | The active pharmaceutical ingredient (API) to be encapsulated. |
| Dialysis Tubing or Purification Columns | For purifying formed nanoparticles from free drugs and solvents. |
| Dynamic Light Scattering (DLS) Instrument | For measuring nanoparticle hydrodynamic size and Polydispersity Index (PDI) [8]. |
| Ultracentrifuge | For separating nanoparticles from the suspension for further analysis. |
| HPLC or UV-Vis Spectrophotometer | For quantifying drug loading and encapsulation efficiency [8]. |
Step-by-Step Procedure
Formulation Variation: Systematically vary one or two critical formulation parameters at a time while holding others constant. Key parameters to vary include:
- Polymer concentration
- Drug-to-polymer ratio
- Aqueous-to-organic phase volume ratio
- Surfactant type and concentration
- Homogenization speed and time
Nanoparticle Synthesis: For each unique formulation, synthesize nanoparticles using a standard method such as single or double emulsion-solvent evaporation.
Purification: Purify the nanoparticle suspension via dialysis or centrifugation to remove the organic solvent and unencapsulated drug.
Characterization and Analysis: For each batch, characterize the nanoparticles by:
- Size and PDI: Measure using Dynamic Light Scattering (DLS) [8].
- Drug Loading (DL) and Encapsulation Efficiency (EE): Determine by lysing a known amount of purified nanoparticles and quantifying the drug content using a validated analytical method (e.g., HPLC). Calculate using [8]:
Drug Loading (%) = (Weight of drug in nanoparticles / Weight of total nanoparticle) x 100Encapsulation Efficiency (%) = (Weight of drug in nanoparticle / Initial weight of blank drug) x 100
Data Compilation and Iteration: Compile the data from all formulations. Analyze results to identify trends. Based on the outcomes, design a new set of formulations for the next round of iterative testing, attempting to converge on the optimal parameters.
Limitations in Practice
This one- or two-parameter-at-a-time approach is inherently inefficient. It fails to capture complex, non-linear interactions between three or more parameters. For instance, as shown in Table 1, the relationship between nanoparticle size, PDI, and encapsulation efficiency for PLGA 50:50 is highly non-linear, with efficiency fluctuating significantly across different sizes and PDIs [8]. Discovering these complex relationships through trial-and-error is a major contributor to the combinatorial challenge, consuming significant time and resources.
A Paradigm Shift: Machine Learning Accelerated Genetic Algorithms (MLaGA)
The core innovation that addresses the combinatorial challenge is the integration of a machine learning model directly into the optimization workflow. This creates a Machine Learning Accelerated Genetic Algorithm (MLaGA), which combines the robust, global search capabilities of a GA with the rapid predictive power of ML [7].
How MLaGA Works
The MLaGA operates with two tiers of energy evaluation: one by the ML model (a surrogate) providing a predicted fitness, and the other by the high-fidelity energy calculator (e.g., DFT) providing the actual fitness [7]. A key implementation uses a nested GA to search the surrogate model, acting as a high-throughput screening function that runs within the "master" GA. This allows the algorithm to make large steps on the potential energy surface without performing expensive energy evaluations [7].
Diagram 1: MLaGA workflow reduces costly calculations. The algorithm cycles between rapid ML-guided search and high-fidelity validation, filtering the vast search space before committing to expensive computations.
Experimental Protocol: Implementing an MLaGA for Nanoparticle Discovery
This protocol details the steps for setting up an MLaGA search, using the example of finding the lowest-energy chemical ordering in a nanoalloy.
Materials and Computational Tools
- Genetic Algorithm Framework: Software capable of performing crossover, mutation, and selection operations on a population of candidate structures [7].
- Machine Learning Model: A probabilistic model, such as a Gaussian Process (GP), to act as the surrogate fitness evaluator [7] [8]. The GP defines a prior over functions and, after observing data, provides a posterior prediction for new input candidates, including uncertainty estimates [8].
- High-Fidelity Energy Calculator: A method like Density Functional Theory (DFT) for accurate but expensive energy evaluations [7].
- Descriptor Generation: A method to convert the atomic structure of a nanoparticle into a numerical vector (descriptor) that the ML model can process.
Step-by-Step Procedure
Initialization:
- Generate an initial population of candidate nanoparticle structures with random chemical ordering.
- Perform a high-fidelity energy calculation (e.g., DFT) on this small initial set to create a seed dataset.
ML Model Training:
- Train the Gaussian Process (or other ML) model on the current dataset of structures and their calculated energies.
Nested Surrogate Search:
- Launch a nested genetic algorithm where the fitness of candidates is evaluated using the trained ML model, not the expensive calculator.
- Run the nested GA for multiple generations, rapidly exploring the search space based on predicted fitness.
High-Fidelity Validation:
- Select the final population from the nested GA and evaluate their energies using the high-fidelity DFT calculator.
Data Augmentation and Iteration:
- Add the new candidates and their validated energies to the training dataset.
- Retrain the ML model on the augmented dataset.
- Repeat steps 3-5 until a convergence criterion is met (e.g., the ML search can no longer find new candidates predicted to be significantly better).
Quantifiable Efficacy of MLaGA
The MLaGA methodology provides a dramatic reduction in computational cost. As shown in Table 1, it can locate the full convex hull of minima using only ~280-1,200 energy calculations, a reduction of over 50-fold compared to the traditional GA [7] [9]. This makes searching through the space of all homotops and compositions of a binary alloy particle feasible using accurate but expensive methods like DFT [7].
This approach is not limited to metallic nanoalloys. The Gaussian Process model has also been successfully applied to predict the properties of polymer nanoparticles like PLGA, generating graphs that predict drug loading and encapsulation efficiency based on nanoparticle size and PDI, thereby eliminating the need for extensive trial-and-error experimentation [8].
The discovery and optimization of novel materials, particularly nanoparticles for drug delivery, represent a critical frontier in advancing human health. However, the computational cost of accurately evaluating candidate materials often renders traditional search methods impractical. Within this challenge lies a powerful synergy: Machine Learning (ML) surrogate models are revolutionizing the efficiency of Genetic Algorithms (GAs), creating a feedback loop that dramatically accelerates computational discovery. This paradigm, known as the Machine Learning accelerated Genetic Algorithm (MLaGA), transforms the discovery workflow. By replacing expensive physics-based energy calculations with a fast-learned model during the GA's search process, the MLaGA framework achieves orders-of-magnitude reduction in computational cost, making previously infeasible searches through vast material spaces not only possible but efficient [7]. This Application Note details the quantitative benchmarks, core protocols, and essential tools for deploying MLaGA in nanoparticle discovery research.
Quantitative Performance Benchmarks
The integration of ML surrogates within a GA framework leads to dramatic improvements in computational efficiency, as quantified by benchmark studies on nanoparticle optimization.
Table 1: Performance Comparison of Traditional GA vs. MLaGA for Nanoparticle Discovery
| Algorithm Type | Number of Energy Calculations | Reduction Factor | Key Features | Reported Search Context |
|---|---|---|---|---|
| Traditional GA | ~16,000 | 1x (Baseline) | Direct energy evaluation for every candidate; robust but slow [7]. | Searching for stable PtxAu147-x nanoalloys across all compositions [7]. |
| MLaGA (Generational) | ~1,200 | ~13x | Uses an on-the-fly trained ML model as a surrogate for a full generation of candidates [7]. | Same as above, using a Gaussian Process model [7]. |
| MLaGA (Pool-based) | ~310 | ~50x | A new ML model is trained after each new energy calculation, maximizing information gain [7]. | Same as above, with serial evaluation [7]. |
| MLaGA (Uncertainty-Aware) | ~280 | ~57x | Incorporates prediction uncertainty into the selection criteria, guiding exploration [7]. | Same as above, using the cumulative distribution function for fitness [7]. |
Experimental Protocols
This section provides a detailed, step-by-step methodology for implementing an MLaGA to discover optimized nanoparticle alloys, as validated in recent literature.
Protocol: MLaGA for Nanoalloy Catalyst Discovery
Application: Identification of stable, compositionally variant Pt-Au nanoparticle alloys for catalytic applications. Primary Objective: To locate the convex hull of stable minima for a 147-atom Mackay icosahedral template structure across all PtxAu147−x (x ∈ [1, 146]) compositions with a minimal number of Density Functional Theory (DFT) calculations [7].
Materials and Data Requirements:
- Initial Dataset: A small, initial population of nanoparticle candidates with known compositions and atomic arrangements (genotypes) and their associated fitness (e.g., energy calculated via DFT or a cheaper potential like EMT).
- Fitness Evaluator: A high-fidelity, computationally expensive energy calculator (e.g., DFT) used sparingly.
- ML Framework: A regression model capable of learning from sequential data (e.g., Gaussian Process Regression, Neural Networks).
- GA Engine: A custom or library-based GA capable of supporting nested or pool-based populations.
Procedure:
Initialization:
- a. Define the search space: a 147-atom binary alloy with a fixed icosahedral geometry but variable chemical ordering and composition [7].
- b. Generate an initial population of candidate nanoparticles with random chemical ordering.
- c. Evaluate the fitness (excess energy) of each candidate in the initial population using the high-fidelity DFT calculator.
ML Surrogate Model Training:
- a. Train an initial ML surrogate model (e.g., Gaussian Process) on the current dataset of nanoparticle genotypes and their corresponding DFT-validated energies [7].
- b. The model learns the complex mapping between the atomic configuration (feature space) and the resulting energy (fitness).
Nested Surrogate GA Search:
- a. Within the main ("master") GA, initiate a nested GA.
- b. The nested GA uses the trained ML surrogate as its fitness function, which is computationally cheap to evaluate.
- c. Run the nested GA for multiple generations. In this phase, candidate offspring are generated via crossover and mutation, and their fitness is predicted by the ML model without performing any new DFT calculations [7].
- d. This step acts as a high-throughput screening, exploring the potential energy surface broadly and cheaply.
Candidate Selection and Validation:
- a. After the nested GA concludes, select the final population of unrelaxed candidates predicted by the surrogate to be the most fit.
- b. Pass these top candidates to the master GA.
- c. Evaluate these candidates using the high-fidelity DFT calculator to obtain their true fitness.
- d. Add these new genotype-true fitness pairs to the growing dataset.
Model Retraining and Iteration:
- a. Retrain the ML surrogate model on the updated, enlarged dataset.
- b. The model's accuracy improves as it learns from more DFT-validated data.
- c. Repeat steps 3-5 until a convergence criterion is met. Convergence is typically signaled when the ML surrogate can no longer find new candidates predicted to be significantly better than the existing best, indicating the search has stabilized [7].
Advanced Protocol: Uncertainty-Aware Pool-Based MLaGA
For maximum efficiency in serial computation, the following variant can be implemented.
Modifications to Base Protocol:
- Population Model: Switch from a generational population to a pool-based population.
- Sequential Training: Train a new ML surrogate model after every single new DFT calculation.
- Uncertainty Exploitation: Use the ML model's predictive uncertainty (e.g., from a Gaussian Process) in the fitness function. A common strategy is to maximize the Expected Improvement (EI) or use the Cumulative Distribution Function of the prediction, which balances exploration (probing uncertain regions) with exploitation (refining known good regions) [7]. This approach can reduce the required energy calculations to approximately 280, a 57-fold improvement over the traditional GA [7].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Computational and Experimental Tools for MLaGA-driven Nanoparticle Research
| Reagent / Tool | Type | Function in MLaGA Workflow | Application Context |
|---|---|---|---|
| Density Functional Theory (DFT) | Computational Calculator | Provides high-fidelity, quantum-mechanical energy and property evaluation for training the ML surrogate and validating final candidates [7]. | Gold-standard for accurate nanoalloy stability and catalytic property prediction [7]. |
| Gaussian Process (GP) Regression | Machine Learning Model | Serves as the surrogate model; provides fast fitness predictions and, crucially, quantifies prediction uncertainty [7]. | Ideal for data-efficient learning in early stages of MLaGA search [7]. |
| Poly(lactide-co-glycolide) (PLGA) | Nanoparticle Polymer | A biodegradable and biocompatible polymer used to form nanoparticle drug delivery carriers [10] [11]. | A key material for formulating nanoparticles designed to deliver small-molecule drugs across biological barriers [11]. |
| Liposomes | Nanoparticle Lipid Vesicle | Spherical vesicles with aqueous cores, used for encapsulating and delivering therapeutic drugs or genetic materials [10] [12]. | Basis for FDA-approved formulations (e.g., Doxil) and modern mRNA vaccine delivery (LNPs) [12]. |
| Mass Photometry | Analytical Instrument | A single-particle characterization technique that measures the molecular mass of individual nanoparticles, revealing heterogeneity in drug loading or surface coating [12]. | Critical quality control for ensuring batch-to-batch consistency of targeted nanoparticle formulations [12]. |
The integration of machine learning surrogate models with genetic algorithms represents a transformative advancement in computational materials discovery. The MLaGA framework, as detailed in these protocols and benchmarks, enables researchers to navigate the immense complexity of nanoparticle design spaces with unprecedented efficiency, reducing the number of required expensive energy evaluations by over 50-fold [7]. This synergy between robust evolutionary search and rapid machine learning prediction creates a powerful, adaptive feedback loop. For researchers in drug development and nanomedicine, adopting the MLaGA approach and the associated toolkit—from uncertainty-aware ML models to single-particle characterization techniques—provides a tangible path to accelerate the rational design of next-generation nanoparticles, ultimately shortening the timeline from conceptual design to clinical application.
The discovery of novel nanomaterials, such as nanoalloy catalysts, is often impeded by the immense computational cost associated with exploring their vast structural and compositional landscape. Genetic algorithms (GAs) are robust metaheuristics for this global optimization challenge, but their requirement for thousands of energy evaluations using quantum mechanical methods like Density Functional Theory (DFT) renders comprehensive searches impractical [7]. This application note details a proof-of-concept case study, framed within a broader thesis on Machine Learning Accelerated Genetic Algorithms (MLaGA), which successfully demonstrated a 50-fold reduction in the number of required energy calculations. By integrating an on-the-fly trained machine learning model as a surrogate for the potential energy surface, the MLaGA methodology makes exhaustive nanomaterial discovery searches feasible, significantly accelerating research and development timelines [7].
Key Performance Metrics
The following table summarizes the quantitative results from benchmarking the MLaGA approach against a traditional genetic algorithm for a specific computational challenge: identifying the convex hull of stable minima for all compositions of a 147-atom PtxAu147−x icosahedral nanoparticle. The total number of possible atomic arrangements (homotops) for this system is approximately 1.78 × 10^44, illustrating the scale of the search space [7].
Table 1: Comparison of Computational Efficiency Between Traditional GA and MLaGA Variants
| Algorithm / Method | Approximate Number of Energy Calculations | Computational Reduction (Compared to Traditional GA) |
|---|---|---|
| Traditional "Brute Force" GA | ~16,000 | 1x (Baseline) |
| MLaGA (Generational Population) | ~1,200 | ~13-fold |
| MLaGA with Tournament Acceptance | <600 | >26-fold |
| MLaGA (Pool-based with Uncertainty) | ~280 | >57-fold |
The data shows a clear hierarchy of efficiency, with the most sophisticated MLaGA implementation, which uses a pool-based population and leverages the prediction uncertainty of the machine learning model, achieving a reduction of more than 57-fold, surpassing the 50-fold target [7].
Experimental Protocols & Workflows
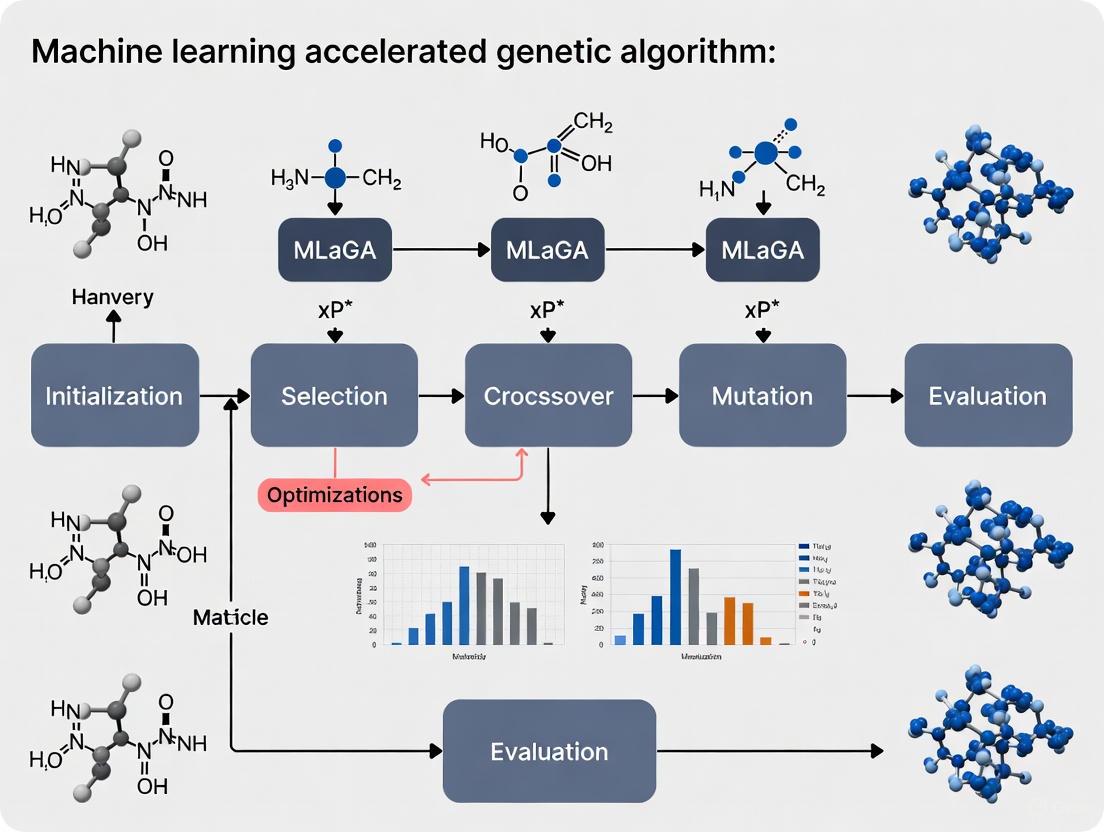
The Machine Learning Accelerated Genetic Algorithm (MLaGA) Workflow
The core innovation of the MLaGA is its two-tiered evaluation system, which combines the robust search capabilities of a GA with the rapid screening power of a machine learning model. The general workflow is illustrated below.
Diagram 1: MLaGA workflow integrating machine learning surrogate model for accelerated discovery.
Protocol 1: Traditional GA Baseline [7]
- Initialization: Generate a population of random candidate nanoparticle structures.
- Fitness Evaluation: Relax each candidate structure and compute its energy using a quantum mechanical calculator (e.g., DFT). This is the most computationally expensive step.
- Selection: Select parent structures based on their fitness (lower energy is better).
- Genetic Operations: Create a new generation of candidates by applying crossover (combining parts of two parents) and mutation (random perturbations) operators.
- Iteration: Repeat steps 2-4 for many generations until the population converges on the lowest-energy (global minimum) structure.
- Output: The algorithm requires ~16,000 energy calculations to map the full convex hull for the benchmark system.
Protocol 2: MLaGA with Generational Population [7]
- Initialization & Initial DFT: Initialize a population and evaluate a small set of candidates with DFT to create an initial training set.
- ML Model Training: Train a Gaussian Process (GP) regression model on the collected DFT data. This model learns to predict the energy of a structure based on its features.
- Nested Surrogate GA: A "nested" GA runs for multiple generations using the ML model as the fitness evaluator. This allows for rapid exploration of the potential energy surface without DFT calculations.
- Injection to Master GA: The best candidates from the nested GA are passed to the "master" GA.
- DFT Validation & Model Update: These candidates are evaluated with DFT for validation, and the new data is used to retrain and improve the ML model.
- Convergence: The process repeats until the ML model can no longer find new candidates predicted to be better than the existing ones. This approach reduces the required DFT calculations to ~1,200.
Protocol 3: Advanced MLaGA with Pool-based Active Learning [7] [13]
- Serial Execution: The GA progresses one candidate at a time in a serial, pool-based manner.
- Per-Candidate Model Update: The ML model is updated after every single DFT calculation, making it highly adaptive.
- Uncertainty Quantification: The GP model provides an uncertainty estimate (variance) for its predictions alongside the predicted energy (mean).
- Informed Selection: The next candidate for DFT evaluation is chosen based on a function that balances predicted energy (exploitation) and prediction uncertainty (exploration). A common method is to use the Upper Confidence Bound (UCB) or the cumulative distribution function.
- Output: This most efficient protocol locates the convex hull in approximately 280-310 DFT calculations.
The Active Learning Loop for On-the-Fly Training
A critical component of the advanced MLaGA is the active learning loop, which ensures the machine learning model is trained on the most informative data points. This workflow is detailed below.
Diagram 2: Active learning loop for on-the-fly training of machine learning potential.
Protocol 4: On-the-Fly Active Learning for Geometry Relaxation [13]
This protocol is used within tools like Cluster-MLP to accelerate the individual geometry relaxation steps in the GA.
- Initial DFT Query: For a new candidate cluster geometry, a single-point DFT calculation is performed to obtain initial energy and forces.
- ML Potential Initialization: This data is used to initialize a machine learning potential (e.g., FLARE++).
- ML-Guided Relaxation: The atomic configuration is updated using an optimizer (e.g., BFGS) driven by the forces predicted by the ML potential.
- Uncertainty Monitoring: The ML potential provides the mean force prediction and the associated uncertainty for each atom.
- Decision Point:
- If the maximum uncertainty of the force prediction is above a set threshold, a new DFT single-point calculation is triggered at the current geometry. This new data is used to retrain the ML potential, improving its accuracy in that region of the potential energy surface.
- If the uncertainty is low and the relaxation is converged (based on ML-predicted forces), the final structure is accepted.
- Output: The fully relaxed structure, verified by DFT at key points, is added to the GA population. This method drastically reduces the number of DFT steps required for each relaxation trajectory.
The Scientist's Toolkit: Essential Research Reagents & Computational Solutions
This section outlines the key software, algorithms, and computational methods that form the essential "toolkit" for implementing an MLaGA for nanomaterial discovery.
Table 2: Key Research Reagent Solutions for MLaGA Implementation
| Tool / Solution | Type | Function in MLaGA Protocol |
|---|---|---|
| Genetic Algorithm (GA) | Metaheuristic Algorithm | Core global search engine for exploring nanocluster configurations via selection, crossover, and mutation [7] [13]. |
| Density Functional Theory (DFT) | Quantum Mechanical Calculator | Provides accurate, reference-quality energy and forces for training the ML model and validating key candidates; the computational bottleneck [7] [13]. |
| Gaussian Process (GP) Regression | Machine Learning Model | Serves as the surrogate energy predictor; chosen for its ability to provide uncertainty estimates alongside predictions [7] [14]. |
| FLARE++ ML Potential | Machine Learning Potential | An interatomic potential used in active learning frameworks for on-the-fly force prediction and uncertainty quantification during geometry relaxation [13]. |
| Birmingham Parallel GA (BPGA) | Genetic Algorithm Code | A specific GA implementation offering diverse mutation operations, modified and integrated into frameworks like DEAP for cluster searches [13]. |
| DEAP (Distributed Evolutionary Algorithms in Python) | Computational Framework | Provides a flexible Python toolkit for rapid prototyping and implementation of evolutionary algorithms, including GAs [13]. |
| ASE (Atomistic Simulation Environment) | Python Library | Interfaces with electronic structure codes and force fields, simplifying the setup and analysis of atomistic simulations within the GA workflow [13]. |
| Active Learning (AL) | Machine Learning Strategy | Manages on-the-fly training of the ML model by strategically querying DFT calculations only when necessary, maximizing data efficiency [13]. |
This proof-of-concept case study establishes the MLaGA framework as a transformative methodology for computational materials discovery. By achieving a 50-fold reduction in the number of required DFT calculations—from ~16,000 to ~280—the approach overcomes a critical bottleneck in the unbiased search for stable nanoclusters and nanoalloys [7]. The detailed protocols for generational and pool-based MLaGA, complemented by active learning for geometry relaxation, provide a clear roadmap for researchers. Integrating robust genetic algorithms with efficient, uncertainty-aware machine learning models renders previously intractable searches feasible, paving the way for the accelerated design of next-generation nanomaterials, such as high-performance nanoalloy catalysts [7] [13].
The discovery and optimization of novel nanoparticles represent a significant challenge in materials science and drug development. The process requires navigating complex, high-dimensional design spaces where evaluations using traditional experimental methods or high-fidelity simulations are prohibitively time-consuming and expensive. This application note details key methodologies—Surrogate-Assisted Evolutionary Algorithms (SAEAs) and Multi-Objective Optimization (MOO)—that, when integrated as a Machine Learning Accelerated Genetic Algorithm (MLaGA), dramatically enhance the efficiency of computational nanoparticle discovery research. We frame these concepts within a practical workflow, provide structured quantitative comparisons, and outline detailed experimental protocols for researchers.
Core Terminology and Concepts
Surrogate Models in Optimization
A surrogate model (also known as a metamodel or emulator) is an engineering method used when an outcome of interest cannot be easily measured or computed directly. It is a computationally inexpensive approximation of a computationally expensive simulation or experiment [15]. In the context of MLaGA for nanoparticle discovery:
- Purpose: To mimic the behavior of a high-fidelity model (e.g., Density Functional Theory (DFT) calculations of nanoparticle energy) as closely as possible while being cheaper to evaluate [15].
- Function: It acts as a surrogate fitness evaluator within a Genetic Algorithm (GA), drastically reducing the number of expensive fitness evaluations required [16] [17]. One study on Pt-Au nanoparticles reported a 50-fold reduction in required energy calculations using this approach [16].
Surrogate models can be broadly classified into two categories used in Single-Objective SAEAs [18]:
- Absolute Fitness Models: Directly approximate the exact fitness value of candidate solutions.
- Relative Fitness Models: Estimate the relative rank or preference of candidates rather than their absolute fitness values.
Multi-Objective Optimization (MOO)
Multi-Objective Optimization is an area of multiple-criteria decision-making concerned with mathematical optimization problems involving more than one objective function to be optimized simultaneously [19]. In nanoparticle design, conflicting objectives are common, such as maximizing catalytic activity while minimizing cost or material usage.
Key MOO concepts include:
- Pareto Optimality: A solution is Pareto optimal if no objective can be improved without degrading at least one other objective [19] [20]. A solution is dominated if another solution exists that is better in at least one objective and at least as good in all others [20].
- Pareto Frontier: The set of all Pareto optimal solutions, representing the optimal trade-offs between the conflicting objectives [19] [21].
Quantitative Data and Model Comparison
Performance of Surrogate Models in SAEAs
The selection of a surrogate model involves a critical trade-off between computational cost and predictive accuracy. The following table summarizes the characteristics of common surrogate types used in SAEAs, based on empirical comparisons [18] [17].
Table 1: Comparison of Surrogate Model Characteristics for SAEAs
| Surrogate Model Type | Computational Cost | Typical Accuracy | Key Strengths | Considerations for Nanoparticle Research |
|---|---|---|---|---|
| Polynomial Response Surfaces | Very Low | Medium | Simple, fast to build and evaluate | May be insufficient for complex, rugged energy landscapes |
| Kriging / Gaussian Processes | High | High | Provides uncertainty estimates, good for adaptive sampling | High cost may negate benefits for very cheap surrogates |
| Radial Basis Functions (RBF) | Low to Medium | Medium to High | Good balance of accuracy and speed | A common and versatile choice for initial implementations |
| Support Vector Machines (SVM) | Medium (Training) | Medium to High | Effective for high-dimensional spaces | Ranking SVMs can preserve optimizer invariance |
| Artificial Neural Networks (ANN) | High (Training) | Very High | High flexibility and accuracy for complex data | Requires large training sets, risk of overfitting |
| Physical Force Fields (e.g., AMOEBA, GAFF) | Medium | Variable (System-Dependent) | Incorporates physical knowledge | Accuracy can be system-dependent; requires careful validation [17] |
MLaGA Efficiency Gains
The core benefit of integrating a surrogate model is a dramatic reduction in computational resource consumption.
Table 2: Documented Efficiency Gains from MLaGA Applications
| Study Context | Traditional Method Cost | MLaGA Cost | Efficiency Gain | Key Surrogate Model |
|---|---|---|---|---|
| Atomic Distribution of Pt-Au Nanoparticles [16] | ~X Expensive Energy Calculations | 50x fewer calculations | 50-fold reduction | Machine Learning Model (unspecified) |
| Peptide Structure Search (GPGG, Gramicidin S) [17] | Months of DFT-MD Simulation | A few hours for GA search + DFT refinement | Reduction from months to hours | Polarizable Force Field (AMOEBApro13) |
Experimental Protocols and Workflows
Protocol: Surrogate-Assisted Genetic Algorithm for Nanoparticle Discovery
This protocol outlines the steps for using an MLaGA to identify low-energy nanoparticle configurations, adapted from successful applications in peptide structure search [17].
1. Objective Definition: * Define the primary objective, e.g., find the atomic configuration of a Pt-Ligand nanoparticle that minimizes the system's potential energy.
2. Initial Sampling and Surrogate Training: * Design of Experiments (DoE): Use a sampling technique (e.g., Latin Hypercube Sampling) to select an initial set of nanoparticle configurations (50-200 individuals) across the design space. * High-Fidelity Evaluation: Run the expensive, high-fidelity simulation (e.g., DFT) on this initial population to obtain accurate fitness values (energy). * Surrogate Model Construction: Train a chosen surrogate model (e.g., a polarizable force field or an RBF network) on the input-output data (nanoparticle configuration -> energy) from the initial sample.
3. Iterative MLaGA Loop: * GA Search with Surrogate: Run a standard Genetic Algorithm (with selection, crossover, and mutation). The surrogate model, not the high-fidelity simulation, is used to evaluate the fitness of candidate solutions. * Infill Selection: From the GA population, select a small subset (e.g., 5-10) of the most promising candidates based on surrogate-predicted fitness (and uncertainty, if available). * High-Fidelity Validation & Update: Evaluate the selected candidates using the high-fidelity simulator (DFT). * Database Update: Add the new [configuration, true fitness] data pairs to the training database. * Surrogate Model Update: Re-train the surrogate model on the enlarged database to improve its accuracy for the next iteration. * Convergence Check: Repeat steps a-d until a convergence criterion is met (e.g., a maximum number of iterations, no improvement in best fitness for several generations, or a target fitness is reached).
4. Final Analysis: * The best-performing configurations validated by high-fidelity simulation are the final output of the optimization.
Protocol: Multi-Objective Optimization for Nanoparticle Design
This protocol describes how to generate a Pareto frontier for a multi-objective problem, such as designing a nanoparticle for both high efficacy and low cytotoxicity.
1. Objective Definition:
* Define the conflicting objectives. For a drug delivery nanoparticle:
* Objective 1: Maximize Drug Loading Capacity (f_load).
* Objective 2: Minimize Predicted Cytotoxicity (f_tox).
2. Method Selection and Implementation (Weighted Sum Method):
* Aggregate Objective: Combine the multiple objectives into a single scalar objective function:
F_obj = w1 * (f_load / f_load0) - w2 * (f_tox / f_tox0)
where w1 and w2 are weighting coefficients ( w1 + w2 = 1), and f_load0, f_tox0 are scaling factors to normalize the objectives to similar magnitudes [21].
* Systematic Weight Variation: Perform a series of single-objective optimizations (using a GA or MLaGA) where the weights (w1, w2) are systematically varied (e.g., (1.0, 0.0), (0.8, 0.2), ..., (0.0, 1.0)).
* Solution Collection: Each optimization run with a unique weight vector will yield one (or a few) Pareto optimal solution(s). Collect all non-dominated solutions from all runs.
3. Post-Processing and Decision Making:
* Construct Pareto Frontier: Plot the objective values of all collected non-dominated solutions. This scatter plot represents the Pareto frontier.
* Trade-off Analysis: Analyze the frontier to understand the trade-offs. For example, how much must f_tox increase to achieve a unit gain in f_load?
* Final Selection: Use domain expertise or higher-level criteria to select the single best-compromise solution from the Pareto frontier.
Visualizing the MLaGA Workflow for Nanoparticle Discovery
The following diagram illustrates the iterative interaction between the genetic algorithm, the surrogate model, and the high-fidelity simulator.
Visualizing Multi-Objective Optimization and Pareto Concepts
This diagram clarifies the core concepts of Pareto optimality and the structure of a multi-objective optimization process.
The Scientist's Toolkit: Essential Research Reagents & Materials
For researchers implementing an MLaGA pipeline for nanoparticle discovery, the following computational and material "reagents" are essential.
Table 3: Essential Research Reagents and Materials for MLaGA-driven Nanoparticle Discovery
| Category | Item / Software / Method | Function / Purpose in the Workflow |
|---|---|---|
| Computational Environments | Python (with libraries like NumPy, Scipy) / Julia | Primary programming languages for implementing the GA, surrogate models, and workflow automation. |
| Surrogate Modeling Libraries | Surrogate Modeling Toolbox (SMT) [15] / Surrogates.jl [15] | Provides a library of pre-implemented surrogate modeling methods (e.g., Kriging, RBF) for easy integration and benchmarking. |
| High-Fidelity Simulators | Density Functional Theory (DFT) Codes (e.g., VASP, Gaussian) [17] | Provides the "ground truth" evaluation of nanoparticle properties (e.g., energy, stability) for training and validating the surrogate model. |
| Approximate Physical Models | Classical Force Fields (e.g., GAFF, AMOEBA) [17] / Semi-Empirical Methods (e.g., PM6, DFTB) | Serves as a physics-informed, medium-accuracy surrogate to pre-screen candidates before final DFT validation. |
| Optimization Algorithms | Genetic Algorithm / Evolutionary Algorithm Libraries | Provides the core search engine for exploring the configuration space of nanoparticles. |
| Nanoparticle Characterization | Scanning/Transmission Electron Microscopy (SEM/TEM) [22] [23] | Used for experimental validation of predicted nanoparticle size, shape, and morphology. |
| Nanoparticle Synthesis | Bottom-Up Synthesis (e.g., Chemical Vapor Deposition) [22] [24] | Physical methods to synthesize the computationally predicted optimal nanoparticle structures. |
From Code to Cure: Implementing MLaGA Workflows for Real-World Nanoparticle Design
The discovery and optimization of functional nanomaterials, such as nanoparticle (NP) alloys, are impeded by the vastness of the compositional and structural search space. Conventional methods, like density functional theory (DFT), are computationally prohibitive for exhaustive exploration. This Application Note details a integrated experimental-computational pipeline that synergizes machine learning-accelerated genetic algorithms (MLaGA), droplet-based microfluidics, and high-content imaging (HCI) to establish a high-throughput platform for the discovery of bimetallic nanoalloy catalysts. We present validated protocols and data handling procedures to accelerate materials discovery, with a specific focus on Pt-Au nanoparticle systems.
The convergence of computational materials design and experimental synthesis is pivotal for next-generation material discovery. Genetic algorithms (GAs) are robust metaheuristic optimization algorithms inspired by Darwinian evolution, capable of navigating complex search spaces to find ideal solutions that are difficult to predict a priori [7]. However, their application with accurate energy calculators like DFT has been limited due to computational cost [7].
The machine learning-accelerated genetic algorithm (MLaGA) surmounts this barrier by using a machine learning model, such as Gaussian Process regression, as a surrogate fitness evaluator, leading to a 50-fold reduction in required energy calculations compared to a traditional GA [7] [25] [9]. This computational efficiency enables the feasible discovery of stable, compositionally variant nanoalloys.
This protocol describes the integration of this computational search with two advanced experimental techniques: microfluidics for controlled, high-throughput synthesis and high-content imaging for deep morphological phenotyping. This pipeline creates a closed-loop system for rapid nanoparticle discovery and characterization.
The following diagram illustrates the logical relationships and data flow within the integrated discovery pipeline.
Protocols and Methodologies
Computational Search with MLaGA
The MLaGA protocol is designed to efficiently locate the global minimum energy structure and the convex hull of stable minima for a given nanoparticle composition.
Key Materials & Software
Table 1: Research Reagent Solutions for Computational Search
| Item | Function/Description |
|---|---|
| Genetic Algorithm (GA) Framework | A metaheuristic that performs crossover, mutation, and selection on a population of candidate structures to evolve optimal solutions [7]. |
| Gaussian Process (GP) Regression Model | A machine learning model that acts as a fast, surrogate fitness (energy) evaluator, dramatically reducing the number of expensive energy calculations required [7]. |
| Density Functional Theory (DFT) | The high-fidelity, computationally expensive energy calculator used to validate candidates and train the ML model [7]. |
| Effective-Medium Theory (EMT) | A less accurate, semi-empirical potential used for initial benchmarking and rapid testing of the algorithm [7]. |
Detailed Protocol: MLaGA for Pt-Au Nanoalloys
- Problem Initialization: Define the template structure (e.g., 147-atom Mackay icosahedron) and the compositional range (e.g., PtxAu147-x for (x \in \left[ {1,146} \right])) [7].
- Initial Population Generation: Create a starting population of candidate nanoparticles with random chemical ordering (homotops).
- Fitness Evaluation (Initial): Calculate the excess energy of the initial population using an energy calculator (e.g., EMT or DFT).
- ML Model Training: Train the GP regression model on the initial dataset of structures and their calculated energies.
- Nested Surrogate GA:
- A nested GA performs multiple generations of evolution (crossover, mutation, selection) using the trained ML model as a cheap fitness predictor.
- This step screens thousands of candidates computationally without performing expensive energy calculations.
- Master GA Evaluation: The final population from the nested GA is evaluated with the high-fidelity energy calculator (DFT).
- Population Update & Iteration: The master GA population is updated with the new, fit candidates. Steps 4-6 are repeated until convergence.
- Convergence Criteria: The search is considered converged when the ML surrogate model can no longer find new candidates predicted to be better than the current best, effectively stalling the search [7].
Performance Data
Table 2: MLaGA Performance Benchmark for a 147-Atom Pt-Au Icosahedral Particle
| Search Method | Approx. Number of Energy Calculations | Reduction Factor vs. Traditional GA |
|---|---|---|
| Traditional 'Brute Force' GA | ~16,000 | 1x (Baseline) [7] |
| MLaGA (Generational) | ~1,200 | 13x [7] |
| MLaGA (Pool-based with Uncertainty) | ~280 | 57x [7] |
Microfluidic Synthesis of Nanoparticles
Intelligent microfluidics enables the automated, high-throughput synthesis of candidate nanoparticles identified by the MLaGA with precise control over reaction conditions [26].
Key Materials
- Photolithography Setup: For fabricating high-precision silicon masters with intricate microchannel designs [26].
- Polydimethylsiloxane (PDMS): A biocompatible, transparent elastomer used for rapid prototyping of microfluidic devices via soft lithography [26].
- Metal Precursor Solutions: Chloroplatinic acid (H2PtCl6) and Gold(III) chloride (HAuCl4) solutions in controlled concentrations.
- Reducing Agent Solution: Sodium borohydride (NaBH4) or ascorbic acid.
- Syringe Pumps: For precise control of fluid flow rates.
Detailed Protocol: Droplet-Based Synthesis
- Device Fabrication: Fabricate a PDMS-based droplet microfluidic device using standard soft lithography techniques, featuring flow-focusing geometry for droplet generation [26].
- Solution Preparation: Prepare the aqueous phase containing the metal precursors and the oil phase (continuous phase) containing a surfactant.
- Device Priming: Load the oil phase into the device to prime the channels.
- Droplet Generation: Simultaneously pump the aqueous and oil phases into the device at optimized flow rates. The flow-focusing geometry will break the aqueous stream into monodisperse droplets.
- In-situ Reduction: Introduce the reducing agent solution via a separate inlet to mix with the droplet stream, initiating nanoparticle synthesis within each droplet.
- Collection & Analysis: Collect the emulsion at the outlet and break the droplets to retrieve the synthesized nanoparticles for characterization.
High-Content Imaging and Phenotypic Analysis
High-content imaging (HCI) provides deep morphological phenotyping of synthesized nanoparticles or biological systems affected by them, generating rich data for validation and model retraining [27].
Key Materials
- High-Content Imager: Automated high-resolution microscope system (e.g., confocal or widefield).
- Cell Stains (for biological assays):
- Image Analysis Software: Software capable of segmenting individual cells/particles and extracting morphological features.
Detailed Protocol: Morphological Profiling
- Sample Preparation: For biological susceptibility testing (e.g., Salmonella Typhimurium), expose isolates to the synthesized nanoparticles or antimicrobials over a time course (e.g., 24 hours, sampling every 2 hours) [27].
- Staining: Treat samples with a panel of fluorescent stains to mark different cellular compartments.
- Automated Imaging: Plate samples in multi-well plates and use the HCI system to automatically capture high-resolution images from multiple sites per well.
- Image Analysis & Feature Extraction: Use the analysis software to segment individual cells and extract ~65 morphological, intensity, and texture features (e.g., cell length, area, fluorescence intensity) for each cell [27].
- Data Aggregation: Average the features from all cells in a field of view to create a single datapoint for analysis.
The workflow for HCI data acquisition and analysis is detailed below.
Data Analysis and ML Classification
- Dimensionality Reduction: Use Principal Coordinate Analysis (PCoA) to visualize high-dimensional imaging data and observe clustering by treatment or susceptibility [27].
- Machine Learning Classification: Train a random forest classifier on the aggregated imaging features. The model can learn to predict properties like antimicrobial susceptibility based solely on morphological characteristics, without prior knowledge of the resistance phenotype or direct exposure to the drug [27]. This model can be used to rapidly characterize new samples.
Data Integration and Closing the Loop
The power of this pipeline is realized by integrating data from all three modules. The morphological data from HCI serves as a rapid, high-throughput validation step for the nanoparticles synthesized by the microfluidic platform. This experimental data can be fed back into the MLaGA to refine the surrogate model, constraining the search space with real-world observations and creating an active learning loop that continuously improves the discovery process.
The discovery and optimization of novel nanoparticles represent a formidable challenge in nanomedicine, characterized by vast combinatorial design spaces and complex, often non-linear, structure–function relationships. Traditional trial-and-error approaches are notoriously resource-intensive, time-consuming, and often fail to predict clinical performance [28]. Within this landscape, active learning (AL) has emerged as a powerful machine learning (ML) paradigm to accelerate discovery. By strategically selecting which experiments to perform, AL aims to maximize learning or performance while minimizing the number of costly experimental iterations [29] [30].
The core challenge in applying active learning lies in navigating the exploration-exploitation trade-off. Exploration involves selecting samples to minimize the uncertainty of the surrogate ML model, thereby enhancing its global predictive accuracy. In contrast, exploitation focuses on selecting samples predicted to optimize the target objective function, such as high cellular uptake or a specific plasmonic resonance [29] [30]. Striking the right balance is critical for the efficient navigation of the design space. This application note details protocols and frameworks for implementing active learning, with a specific focus on balancing this trade-off for optimal nanoparticle formulation within a broader thesis investigating Machine Learning Accelerated Genetic Algorithms (MLaGA).
Core Active Learning Framework and the Exploration-Exploitation Trade-off
Active learning operates through an iterative, closed-loop workflow. A surrogate model is initially trained on a small dataset. This model then guides each subsequent cycle by selecting the most informative samples to test next based on an acquisition function. The new experimental results are added to the training set, and the model is updated, creating a continuous feedback loop that refines the understanding of the design space [29] [30].
The acquisition function is the primary mechanism for managing the exploration-exploitation trade-off. The table below summarizes common strategic approaches to this dilemma.
Table 1: Strategic Approaches to the Exploration-Exploitation Trade-off in Active Learning
| Strategy | Core Principle | Typical Use Case |
|---|---|---|
| Exploration-Based | Selects samples where the model's prediction uncertainty is highest. Aims to improve the overall model accuracy [29]. | Early stages of learning or when the design space is poorly understood. |
| Exploitation-Based | Selects samples predicted to have the most desirable property (e.g., highest uptake, target resonance) [29]. | When the primary goal is immediate performance optimization. |
| Balancing Strategies | Explicitly considers both uncertainty and performance to select samples [29]. | The most common approach for robust and efficient optimization. |
| Multi-Objective Optimization (MOO) | Frames exploration and exploitation as explicit, competing objectives and identifies the Pareto-optimal set of solutions, avoiding the bias of a single scalar score [31]. | For complex design spaces where the trade-off is not well-defined; provides a unifying perspective. |
A generic workflow for an active learning-driven discovery platform, integrating the key technological components, is illustrated below.
Application Note: Active Learning for High-Uptake Nanoparticle Design
Integrated Workflow for PLGA-PEG Nanoparticles
Ortiz-Perez et al. (2024) demonstrated a seminal integrated workflow for designing PLGA-PEG nanoparticles with high cellular uptake in human breast cancer cells (MDA-MB-468) [30]. This protocol successfully combines microfluidic formulation, high-content imaging (HCI), and active machine learning into a rapid, one-week experimental cycle.
Table 2: Key Performance Metrics from the PLGA-PEG Active Learning Study [30]
| Metric | Initial Training Set | After Two AL Iterations | Notes |
|---|---|---|---|
| Cellular Uptake (Fold-Change) | ~5-fold | ~15-fold | Measured as normalized fluorescence intensity per cell. |
| Cycle Duration | - | 1 week per iteration | From formulation to next candidate selection. |
| Key Technologies | Microfluidics, HCI, Active ML | Microfluidics, HCI, Active ML | Integrated into a semi-automated platform. |
Detailed Experimental Protocol
Objective: To identify a PLGA-PEG nanoparticle formulation that maximizes cellular uptake using an active learning-guided workflow.
Materials and Reagents
- Polymers: PLGA, PLGA-PEG, PLGA-PEG-COOH, PLGA-PEG-NH₂.
- Solvents: Acetone (or other suitable water-miscible solvent), DiH₂O (anti-solvent).
- Microfluidic Device: Hydrodynamic Flow Focusing (HFF) chip with Y-junction.
- Cell Line: MDA-MB-468 human breast cancer cells.
- Staining Reagents: Hoechst (nuclei), CellMask (membrane), fluorescent dye (e.g., Cy5) for nanoparticle encapsulation.
- Equipment: Programmable syringe pumps, rotary valve, automated fluorescence microscope, CellProfiler software.
Procedure
Initial Library Design & Microfluidic Formulation:
- Define the design space by varying the ratios of the four polymer components and the solvent/anti-solvent Flow Rate Ratio (FRR).
- Use the automated microfluidic setup. The polymer mixtures are loaded into separate reservoirs connected to a rotary valve on the solvent pump. DiH₂O is loaded into the anti-solvent pump.
- For each formulation, the syringe pumps and rotary valve are programmed to mix the specified polymer combination and inject it into the middle channel at a constant flow rate. The anti-solvent flow rate is adjusted to achieve the target FRR, controlling nanoparticle size.
- Encapsulate a fluorescent dye in situ during the nanoprecipitation process to enable subsequent imaging.
High-Content Imaging (HCI) and Analysis:
- Seed MDA-MB-468 cells in a 96-well plate and incubate with the formulated nanoparticles.
- After incubation, fix the cells and stain nuclei and membranes.
- Acquire widefield fluorescence images using an automated microscope with appropriate channels (DAPI for nuclei, Cy5 for nanoparticles, etc.).
- Process images using a CellProfiler pipeline:
- Nuclear Segmentation: Identify individual cells using the nuclei channel.
- Membrane Segmentation: Define cytoplasmic regions using the membrane stain.
- Intensity Quantification: Measure the mean fluorescent intensity from the nanoparticle channel within the segmented cell areas. The distribution of cell intensities is expected to fit a gamma distribution [30].
Active Machine Learning Cycle:
- Model Training: Train a surrogate model (e.g., Gaussian Process Regression, Random Forest) on the current dataset, where the inputs are the formulation parameters (component ratios, FRR) and the output is the measured cellular uptake.
- Candidate Selection: Use an acquisition function on the trained model to select the next set of promising formulations.
- For a balanced approach, use the Upper Confidence Bound (UCB) or Expected Improvement (EI).
- For a more advanced, bias-free approach, implement a Multi-Objective Optimization (MOO) that treats predicted performance (exploitation) and predictive uncertainty (exploration) as separate objectives to identify a Pareto front of optimal candidates [31]. The final candidate can be selected from this front using a method like the knee point or a reliability estimate [31].
- Iteration: The selected formulations are synthesized and tested (return to Step 1), and the data is used to update the model. This loop continues until a stopping criterion is met (e.g., uptake exceeds a target threshold, or performance plateaus).
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Active Learning-Driven Nanoparticle Formulation
| Item | Function / Role in the Workflow |
|---|---|
| PLGA-PEG Copolymer Library | The core building blocks for self-assembling nanoparticles; varying ratios and end-groups (COOH, NH₂) control physicochemical properties like PEGylation, charge, and stability [30]. |
| Hydrodynamic Flow Focusing (HFF) Microfluidic Chip | Enables reproducible, automated, and size-tunable synthesis of highly monodispersed nanoparticles by controlling the solvent/anti-solvent mixing rate [30]. |
| Programmable Syringe Pumps with Rotary Valve | Provides automated and precise fluid handling for high-throughput formulation of different polymer compositions without manual intervention [30]. |
| Fluorescent Dyes (e.g., Cy5) | Used for in situ encapsulation during nanoprecipitation to label nanoparticles, enabling quantification of biological responses like cellular uptake via fluorescence microscopy [30]. |
| Automated Fluorescence Microscope | The core of High-Content Imaging (HCI); allows for rapid, automated acquisition of multiparametric data from cell-based assays in multi-well plates [30]. |
| CellProfiler Software | Open-source bioimage analysis software used to create automated pipelines for segmenting cells and quantifying nanoparticle uptake or other biological responses from HCI data [30]. |
Integration with Genetic Algorithms for MLaGA
The principles of active learning dovetail seamlessly with evolutionary strategies like Genetic Algorithms (GAs), forming a powerful MLaGA framework for nanoparticle discovery. GAs are population-based global optimization algorithms inspired by natural selection, where a population of candidate solutions (e.g., nanoparticle formulations) evolves over generations [32].
In an MLaGA framework, the active learning surrogate model can dramatically accelerate the GA's convergence. Instead of physically testing every individual in a population—a process that is often prohibitively slow and expensive—the ML model can be used to pre-screen and evaluate the fitness of candidate formulations [32]. This allows the algorithm to explore a much larger region of the design space computationally and only validate the most promising individuals experimentally. The experimental results then feedback to retrain and improve the surrogate model, creating a virtuous cycle. This hybrid approach is particularly potent for navigating high-dimensional design spaces where traditional methods struggle [28] [32].
The following diagram illustrates the architecture of this integrated MLaGA system.
Biomedical Rationale and Significance
Poly(lactic-co-glycolic acid)-polyethylene glycol (PLGA-PEG) nanoparticles represent a cornerstone of modern nanomedicine, offering a powerful platform for targeted drug delivery. These nanoparticles synergistically combine the biodegradable, biocompatible, and drug-encapsulating properties of PLGA with the stealth characteristics conferred by PEGylation, which reduces opsonization and recognition by the immune system [33]. This extended circulation time significantly increases the likelihood of nanoparticles reaching their intended target site, a crucial advantage in applications such as cancer therapy where it enables reduced dosage frequency while simultaneously improving therapeutic efficacy [33]. The versatility of PLGA-PEG nanoparticles allows for the encapsulation of a wide spectrum of therapeutic agents, including small molecules, proteins, and nucleic acids, protecting them from degradation and enhancing their absorption [33].
The optimization of these nanoparticles for targeted cellular uptake is paramount for overcoming fundamental challenges in drug delivery, particularly the specific targeting of cancer cells while protecting healthy tissues in conventional chemotherapy [33]. This targeting is achieved through a two-pronged approach: passive and active targeting. The enhanced permeability and retention (EPR) effect facilitates passive accumulation within tumor tissues due to their leaky vasculature and poor lymphatic drainage [34]. More precise active targeting is accomplished by functionalizing the nanoparticle surface with specific ligands, such as antibodies, peptides, or aptamers, that preferentially bind to receptors overexpressed on target cells [33]. This active targeting enhances cellular uptake and intracellular drug release, leading to higher drug concentrations at the disease site and minimized off-target effects [33]. The optimization of nanoparticle properties—such as size, surface charge, PEG density, and ligand orientation—is therefore a critical determinant of their success, creating a complex, multi-variable problem ideally suited for advanced computational optimization methods like Machine Learning accelerated Genetic Algorithms (MLaGA).
Key Characteristics and Optimization Parameters
The performance of PLGA-PEG nanoparticles in targeted drug delivery is governed by a set of interdependent physicochemical properties. These properties must be carefully balanced to achieve optimal systemic circulation, tissue penetration, and cellular uptake. The following table summarizes the core parameters that require optimization.
Table 1: Key Parameters for Optimizing PLGA-PEG Nanoparticles
| Parameter | Optimal Range/Type | Impact on Performance |
|---|---|---|
| Particle Size [33] | 10 - 200 nm | Influences circulation time, tissue penetration, and cellular uptake; smaller particles typically exhibit deeper tissue penetration. |
| Surface Charge (Zeta Potential) [33] | Near-neutral or slightly negative | Reduces non-specific interactions with serum proteins and cell membranes, prolonging circulation time. |
| PEG Molecular Weight & Density [33] | Tunable (e.g., 2k - 5k Da) | Forms a hydrophilic "stealth" corona that minimizes opsonization and clearance by the mononuclear phagocyte system. |
| Drug Encapsulation Efficiency [34] | High (>70-80%) | Determines the total therapeutic payload delivered per nanoparticle, directly impacting efficacy. |
| Drug Release Kinetics [33] | Sustained release (days to weeks) | Controlled by PLGA composition (lactide:glycolide ratio) and molecular weight; ensures prolonged therapeutic action. |
| Targeting Ligand Density [33] | Optimized for receptor saturation | Balances specific cellular uptake against potential immune recognition; too high a density can compromise stealth properties. |
Experimental Protocols for Preparation and Characterization
Preparation of PLGA-PEG Nanoparticles via Emulsion Solvent Evaporation
This is a widely used and robust method for synthesizing PEGylated PLGA nanoparticles, particularly for hydrophobic drugs [33].
- Polymer and Drug Solution Preparation: Dissolve the PLGA polymer and the hydrophobic drug in a water-immiscible organic solvent, such as dichloromethane or chloroform [34].
- Aqueous Phase Preparation: Prepare an aqueous solution containing a stabilizer, typically polyvinyl alcohol (PVA) at a concentration of 1-3% [33].
- Emulsification: Add the organic phase to the aqueous phase and emulsify the mixture using a high-speed homogenizer (e.g., 10,000-15,000 rpm for 2-5 minutes) or probe sonication to form a primary oil-in-water (o/w) emulsion [33] [34].
- Solvent Evaporation: Pour the formed emulsion into a larger volume of water under moderate stirring. Stir continuously for 3-4 hours to allow the organic solvent to evaporate, solidifying the nanoparticles [33].
- Purification: Collect the nanoparticles by ultracentrifugation (e.g., 20,000-30,000 rpm for 30 minutes). Wash the pellet with Milli-Q water or distilled water 2-3 times to remove residual solvent and PVA [33].
- Lyophilization: Re-suspend the purified nanoparticles in a cryoprotectant solution (e.g., mannitol or trehalose) and freeze-dry for long-term storage.
Ligand Conjugation for Active Targeting
Active targeting is achieved by conjugating specific ligands (e.g., antibodies, peptides) to the terminal functional group (commonly carboxyl) of the PEG chain [33].
- Ligand Solution: Prepare a solution of the targeting ligand in a suitable buffer, such as phosphate-buffered saline (PBS) at pH 7.4 [33].
- Surface Activation: To the nanoparticle suspension, add a cross-linking agent such as 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) along with N-Hydroxysuccinimide (NHS). Incubate with gentle shaking for 15-30 minutes to activate the carboxyl groups on the nanoparticle surface, forming an NHS ester [33].
- Conjugation Reaction: Add the ligand solution to the activated nanoparticles and allow the reaction to proceed for 2-12 hours at room temperature with gentle agitation.
- Purification: Purify the ligand-anchored nanoparticles via ultracentrifugation or dialysis to remove unreacted cross-linkers and ligands. The final product can be characterized to confirm ligand attachment and density.
Essential Characterization Techniques
Rigorous characterization is critical to link nanoparticle properties to biological performance.
- Size and Size Distribution: Dynamic Light Scattering (DLS) is used to determine the hydrodynamic diameter and polydispersity index (PDI) [33].
- Surface Charge: Zeta potential analysis measures the surface charge, which indicates colloidal stability [33].
- Surface Morphology: Techniques like Scanning Electron Microscopy (SEM) or Transmission Electron Microscopy (TEM) provide visual confirmation of nanoparticle size, shape, and surface texture.
- Drug Encapsulation and Loading: The amount of drug encapsulated is quantified by HPLC or UV-Vis spectroscopy after dissolving the nanoparticles. Encapsulation Efficiency (EE%) is calculated as (Actual drug loading / Theoretical drug loading) × 100%.
- In-vitro Drug Release: Nanoparticles are suspended in a release medium (e.g., PBS at 37°C). At predetermined time points, samples are centrifuged, and the supernatant is analyzed for drug content to build a release profile [33].
Diagram 1: MLaGA-driven optimization workflow for PLGA-PEG nanoparticles. The cycle integrates computational prediction with experimental validation to rapidly converge on an optimal design.
Integration with MLaGA for Accelerated Discovery
The optimization of PLGA-PEG nanoparticles involves navigating a high-dimensional parameter space where interactions between variables are complex and non-linear. A traditional "brute-force" experimental approach is often prohibitively time-consuming and resource-intensive. The MLaGA framework provides a powerful solution to this challenge by combining the robust search capabilities of Genetic Algorithms (GAs) with the predictive power of Machine Learning (ML) [7].
In this paradigm, the physicochemical parameters from Table 1 and the resulting experimental data (e.g., cellular uptake efficiency) form the feature and target spaces for the ML model. The MLaGA workflow, as illustrated in Diagram 1, operates as follows:
- Initialization: An initial population of nanoparticle designs, each defined by a unique combination of parameters (e.g., size, PEG density, ligand type), is generated, either randomly or based on prior knowledge.
- Surrogate Model Training: An ML model (e.g., Gaussian Process Regression) is trained on-the-fly using existing experimental data to act as a computationally inexpensive surrogate for predicting nanoparticle fitness (e.g., targeting efficacy) [7].
- Evolutionary Search: The GA performs selection, crossover, and mutation operations on the population. The fitness of new candidate designs is evaluated using the fast ML surrogate model instead of costly experiments [7].
- Experimental Validation and Feedback: The most promising candidates predicted by the ML model are synthesized and tested experimentally. This new, high-quality data is then fed back into the dataset to retrain and improve the accuracy of the surrogate model [7].
This closed-loop approach leads to a highly efficient search. Benchmark studies have demonstrated that MLaGA can achieve a 50-fold reduction in the number of required energy calculations (or, by analogy, complex experimental evaluations) compared to a traditional GA to locate an optimal solution [7] [16]. This makes it feasible to comprehensively search through vast combinatorial spaces, such as all possible homotops and compositions of a nano-alloy, which would be intractable using conventional methods [7].
Table 2: MLaGA vs. Traditional Workflows for Nanoparticle Optimization
| Aspect | Traditional Empirical Approach | MLaGA-Accelerated Approach |
|---|---|---|
| Experimental Throughput | Low; relies on sequential, one-variable-at-a-time testing. | High; uses ML to pre-screen candidates, focusing experiments only on the most promising leads. |
| Parameter Space Exploration | Limited; practical constraints force a narrow focus. | Extensive; can efficiently explore high-dimensional, combinatorial spaces (e.g., composition, size, ligand). |
| Resource Consumption | High; requires synthesizing and testing a large number of sub-optimal candidates. | Drastically reduced; one study reported a 50-fold reduction in required evaluations [7]. |
| Discovery Timeline | Long and iterative, often taking months to years. | Significantly accelerated; enables rapid convergence to optimal designs in weeks. |
| Insight Generation | Often correlative; limited ability to model complex parameter interactions. | Predictive; the trained ML model can reveal non-linear relationships and design rules. |
The Scientist's Toolkit: Research Reagent Solutions
A successful MLaGA-driven research program relies on high-quality, well-characterized materials and reagents. The following table details essential components for developing and optimizing PLGA-PEG nanoparticles.
Table 3: Essential Research Reagents for PLGA-PEG Nanoparticle Development
| Reagent / Material | Function / Role | Examples / Notes |
|---|---|---|
| PLGA Polymers [33] | Biodegradable copolymer core; encapsulates drug and controls release kinetics. | Varying lactide:glycolide ratios (e.g., 50:50, 75:25) and molecular weights to tune degradation and release profiles. |
| PLGA-PEG Copolymers [33] | Provides the "stealth" matrix; PEG chain extends circulation time and provides a handle for ligand conjugation. | Available with different PEG chain lengths (e.g., 2k, 5k Da) and terminal functional groups (e.g., COOH, NH₂). |
| Stabilizers / Surfactants [33] | Prevents nanoparticle aggregation during synthesis. | Polyvinyl Alcohol (PVA), Poloxamers. Critical for controlling particle size and distribution. |
| Cross-linking Agents [33] | Facilitates covalent attachment of targeting ligands to the nanoparticle surface. | EDC (1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide) and NHS (N-Hydroxysuccinimide) for carboxyl-amine coupling. |
| Targeting Ligands [33] | Enables active targeting by binding to specific receptors on target cells. | Antibodies (e.g., anti-VEGFR2 [34]), peptides (e.g., RGD), aptamers, or small molecules (e.g., folic acid). |
| Characterization Kits | Essential for quantifying success of synthesis and functionalization. | BCA or Bradford assays for protein ligand quantification, Zeta Potential and DLS measurement kits. |
| Cell-Based Assay Kits | Measures the biological efficacy of the optimized nanoparticles. | Cellular uptake assays (e.g., using flow cytometry), cytotoxicity assays (e.g., MTT, MTS). |
Diagram 2: Schematic structure of a targeted PLGA-PEG nanoparticle, showing the drug-loaded core, stealth PEG layer, and surface-conjugated targeting ligand.
The integration of Machine Learning-accelerated Genetic Algorithms (MLaGAs) represents a paradigm shift in the computational design and experimental synthesis of advanced nanomaterials, notably MXenes and gold nanoparticles (Au NPs). This approach synergizes the robust global search capabilities of genetic algorithms (GAs) with the predictive power of machine learning (ML) surrogate models, dramatically accelerating the discovery and optimization process for applications ranging from nanozymes to targeted drug delivery [7] [35]. The table below summarizes the quantitative performance gains and key applications of this methodology.
Table 1: Performance and Applications of MLaGA in Nanomaterial Design
| Metric / Application | Traditional GA | MLaGA | Key Application Example |
|---|---|---|---|
| Number of Energy Calculations | ~16,000 [7] | ~300 (up to 50x reduction) [7] | Searching Pt-Au nanoalloy compositions [7] |
| Search Feasibility | Infeasible for large composition spaces [7] | Feasible with DFT calculations [7] | Identification of core-shell Au-Pt structures [7] |
| Primary ML Role | N/A | On-the-fly surrogate model for energy prediction [7] | Predicting nanoparticle stability and catalytic activity [7] |
| Experimental Functionality | N/A | Guides synthesis of multifunctional composites [36] | Creating Ti3C2Tx-Au cascade nanozymes [36] |
Detailed Experimental Protocols
Protocol 1: MLaGA for Nanoalloy Catalyst Discovery
This protocol outlines the computational discovery of stable bimetallic nanoparticle alloys, such as platinum-gold (Pt-Au), using the MLaGA framework [7].
I. Initialization and First-Principles Calculations
- Population Generation: Initialize a population of candidate nanoparticle structures. For a 147-atom Mackay icosahedral template, this involves defining different chemical orderings (homotops) across compositions PtxAu147−x for x from 1 to 146 [7].
- Fitness Evaluation (First Generation): Use Density Functional Theory (DFT) to calculate the excess energy of each candidate structure in the initial population. This serves as the fitness metric for selection [7].
II. Machine Learning Surrogate Model Training
- Dataset Creation: The DFT-calculated structures and their energies form the initial training dataset.
- Model Selection and Training: Train a Gaussian Process (GP) regression model (or other ML frameworks like deep learning) on this dataset. The model learns to predict the energy of a nanoparticle based on its structural and compositional features, acting as a computationally cheap surrogate for DFT [7].
III. ML-Accelerated Evolutionary Search
- Generational Loop:
- Crossover and Mutation: Apply genetic operators to the parent population to generate a large pool of offspring candidates [7].
- Surrogate Screening: Use the trained GP model to predict the energies of all offspring candidates. Run a nested GA on the surrogate model to identify the most promising candidates predicted to have low energy [7].
- Selective DFT Validation: Perform actual DFT calculations only on the small subset of candidates filtered by the surrogate model. This step validates the ML predictions and provides new, high-quality data [7].
- Model Retraining: Update the GP model with the new DFT-validated data to improve its predictive accuracy for the next generation [7].
- Convergence: The algorithm is considered converged when the ML routine can no longer find new candidates predicted to be better than the existing population, indicating that the global minimum is likely found [7].
Protocol 2: In-Situ Synthesis of MXene-Gold Nanozymes
This experimental protocol details the fabrication of Ti3C2Tx MXene nanosheets loaded with Au NPs (Ti3C2Tx-Au) for enhanced catalase-like and glucose oxidase-like activity, forming a cascade system for tumor therapy [36].
I. Synthesis of Monolayer Ti3C2Tx MXene
- Etching: Begin with bulk Ti3AlC2 MAX phase ceramic. Etch the aluminum (Al) layer using a mixture of lithium fluoride (LiF) and hydrochloric acid (HCl). A typical molar ratio is LiF/HCl (1:6), reacting at 40°C for 24 hours to achieve high delamination efficiency [36] [37].
- Delamination and Ultrasonication: Separate the resulting multilayer Ti3C2Tx and subject it to ultrasonication in an ice bath under a nitrogen (N2) atmosphere to produce small-sized, monolayer MXene nanosheets [36].
II. In-Situ Deposition of Gold Nanoparticles
- Principle: Exploit the inherent reducing nature of Ti3C2Tx MXene, which contains lower valence state Titanium ions [36].
- Procedure: Immerse the prepared Ti3C2Tx MXene nanosheet dispersion in an aqueous solution of chloroauric acid (HAuCl4). The MXene nanosheets spontaneously reduce Au³⁺ ions to Au⁰, leading to the nucleation and growth of Au NPs directly on the MXene surface without requiring an external reducing agent [36].
- Purification: Recover the resulting Ti3C2Tx-Au composite via centrifugation and wash to remove any unreacted precursors or by-products.
III. Surface Functionalization for Biocompatibility
- PEGylation: To enhance stability and suitability for in vivo applications, modify the Ti3C2Tx-Au composite with polyethylene glycol (PEG). This is achieved by incubating the nanocomposite with PEG molecules, which attach to the surface via physical adsorption or covalent bonding, resulting in the final product, TANP (Ti3C2Tx-Au-PEG) [36].
IV. Characterization and Enzymatic Activity Validation
- Material Characterization: Use techniques such as transmission electron microscopy (TEM) to confirm the sheet-like morphology of MXene and the successful loading of Au NPs. X-ray photoelectron spectroscopy (XPS) can verify the reduction of gold and the surface chemistry [36].
- Catalase-like Activity Assay: Monitor the production of O2 from H2O2 decomposition using a dissolved oxygen meter. Compare the reaction rates of Ti3C2Tx-Au against pristine Ti3C2Tx to quantify the enhancement in catalase-like activity [36].
- Glucose Oxidase-like Activity Assay: Measure the consumption of dissolved oxygen or the production of H2O2 and gluconic acid when the nanozyme is incubated with glucose. This confirms the GOx-like property imparted by the Au NPs [36].
Workflow and Pathway Visualization
Diagram 1: MLaGA for nanoparticle discovery.
Diagram 2: Synthesis of MXene-gold nanozymes.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for MLaGA-Guided MXene and Gold Nanoparticle Research
| Reagent/Material | Function and Role in Research | Example/Specification |
|---|---|---|
| MAX Phase Precursor | The starting material for MXene synthesis. Provides the layered structure from which the 'A' element is etched [36] [37]. | Ti3AlC2 ceramic is the most common precursor for Ti3C2Tx MXene [36]. |
| Etching Agents | Selectively removes the 'A' layer from the MAX phase, creating multilayered MXene [36] [37]. | LiF/HCl mixture (e.g., 1:6 molar ratio) is a common, relatively safe etchant [36]. Hydrofluoric Acid (HF) is a more aggressive alternative [37]. |
| Chloroauric Acid (HAuCl4) | The gold precursor for nanoparticle synthesis. It is reduced to metallic gold (Au⁰) to form Au NPs [36]. | Used for in-situ deposition on MXene surfaces due to the reducing capability of Ti3C2Tx [36]. |
| Surface Modifiers | Enhances biocompatibility, colloidal stability in physiological environments, and can impart stealth properties for in vivo applications [36] [38]. | Polyethylene Glycol (PEG) is widely used for PEGylation [36]. Other options include Chitosan and Polyvinylpyrrolidone (PVP) [38]. |
| Density Functional Theory (DFT) | The high-accuracy, computationally expensive energy calculator used to validate the stability and properties of predicted nanostructures, providing ground-truth data for ML training [7]. | Used within the MLaGA loop to calculate the excess energy of nanoparticle homotops [7]. |
The development of Poly(lactic-co-glycolic acid) (PLGA)-based nanoparticles represents a cornerstone of modern pharmaceutical sciences, offering solutions for controlled drug delivery and enhanced therapeutic efficacy. However, the formulation process is inherently complex, with critical quality attributes (CQAs) such as particle size, encapsulation efficiency (E.E.%), and drug loading (D.L.%) being highly sensitive to minor variations in input parameters [39]. A thorough understanding of how polymer properties—specifically molecular weight (Mw) and lactide to glycolide (LA/GA) ratio—interact with process variables is essential for systematic nanocarrier design. This application note decodes these influential parameters within the innovative context of Machine Learning accelerated Genetic Algorithms (MLaGA), providing a structured framework for accelerated nanoparticle discovery and optimization.
Quantitative Analysis of Critical Input Parameters
The relationships between material attributes, process parameters, and the resulting nanoparticle characteristics have been quantitatively analyzed from extensive formulation data. The tables below summarize these critical dependencies.
Table 1: Impact of PLGA Material Attributes on Critical Quality Attributes (CQAs)
| Material Attribute | Impact on Particle Size | Impact on Encapsulation Efficiency (E.E.%) | Impact on Drug Loading (D.L.%) | Influence on Drug Release Kinetics |
|---|---|---|---|---|
| Polymer Molecular Weight (Mw) | Positive correlation; higher Mw generally increases particle size [40] | Influential feature; moderate positive correlation with E.E.% [40] | Less direct impact than LA/GA ratio [40] | Higher Mw leads to slower polymer degradation and more sustained release [39] |
| LA/GA Ratio | Moderate correlation; 50:50 ratio often yields smaller particles [41] [42] | Not the most determining feature [40] | The most determining material attribute for D.L.% [40] | Lower LA content (more hydrophilic) accelerates hydration and degradation, leading to faster release [39] |
| Polymer End Group | Indirect influence via degradation rate | Affects initial burst release and protein interaction [39] | Impacts compatibility with specific drug molecules | Carboxylate end groups accelerate erosion compared to ester end caps [39] |
Table 2: Effect of Critical Process Parameters on Nanoparticle CQAs
| Process Parameter | Impact on Particle Size | Impact on E.E.% and D.L.% | Key Relationships |
|---|---|---|---|
| Drug to Polymer Ratio | Secondary influence compared to solvent and surfactant choices [40] | Strong positive correlation with Loading Capacity (LC) [41] | Fundamental parameter for controlling drug content |
| Surfactant Concentration/Type | Significant impact; determines emulsion stability and droplet size [39] [40] | High E.E.% relies on stable emulsion formation during processing [39] | Hydrophilic-Lipophilic Balance (HLB) is a critical feature [41] |
| Aqueous to Organic Phase Volume Ratio | Key parameter in emulsion-based methods (e.g., nanoprecipitation) [41] | Impacts drug partitioning during formulation | A critical parameter identified via machine learning feature analysis [40] |
| Solvent Polarity | A highly influential parameter on particle size [40] | Affects the diffusion rate of organic solvent, influencing drug trapping [39] | Polarity index is a key descriptor in formulation datasets [41] |
| Homogenization Parameters (Rate, Time) | Directly controls the shear forces and resultant droplet size in emulsions [39] | Affects the integrity of the emulsion, thereby influencing drug leakage | A Critical Processing Parameter (CPP) in emulsion-solvent evaporation [39] [43] |
Experimental Protocols for Parameter Investigation
Protocol 1: Nanoprecipitation for Systematic Formulation Screening
This protocol is ideal for generating high-throughput data for MLaGA training by exploring a wide range of material and process parameters [41] [42].
- Organic Phase Preparation: Dissolve PLGA polymer (varying Mw and LA/GA ratio) and the drug candidate (e.g., Coumarin-6) in a water-miscible organic solvent (e.g., acetone). Systematically vary the drug-to-polymer ratio and polymer concentration [42].
- Aqueous Phase Preparation: Prepare an aqueous solution containing a surfactant (e.g., Poloxamer 188 (Pluronic F68) or Polyvinyl Alcohol (PVA)) at a specified concentration (e.g., 0.04 mM) [42].
- Nanoparticle Formation: Infuse the organic phase into the aqueous phase under controlled stirring or using a microfluidic device. For a continuous process, use a microreactor with set flow rates (e.g., aqueous-to-organic flow rate ratio of 10:1) and a defined residence time (e.g., 5 minutes) [42].
- Solvent Removal & Harvesting: Remove the organic solvent under reduced pressure with moderate stirring. Purify the resulting nanoparticle suspension by repeated centrifugation and washing with Milli-Q water [42].
- Characterization: Analyze the nanoparticles for size, PDI, and zeta potential using Dynamic Light Scattering (DLS). Determine E.E.% and D.L.% using validated analytical methods (e.g., HPLC) [41] [42].
Protocol 2: Quality by Design (QbD) for Parameter Optimization
This protocol uses a systematic Design of Experiments (DoE) approach to model and optimize the formulation process, identifying Critical Process Parameters (CPPs) [43].
- Define Quality Target Product Profile (QTPP): Establish the target CQAs (e.g., particle size < 200 nm, E.E.% > 80%, specific release profile) [39] [43].
- Identify Critical Material Attributes (CMAs) and CPPs: Select input variables for screening (e.g., PLGA Mw, LA/GA ratio, surfactant concentration, homogenization speed, aqueous-to-organic phase volume ratio) [43].
- Design of Experiments (DoE): Utilize a Response Surface Methodology (RSM) design to create an experimental matrix that efficiently explores the design space, including interaction and quadratic effects between parameters [43].
- Fabrication and Analysis: Execute the DoE matrix by fabricating batches of nanoparticles (e.g., via double-emulsion) and characterize all CQAs for each batch [43].
- Model Building and Optimization: Build mathematical models correlating CMAs/CPPs to CQAs. Identify the design space where the CQAs meet the pre-defined QTPP criteria [43].
Visualizing the MLaGA-Driven Discovery Workflow
The following diagrams illustrate the integrated experimental and computational workflow for nanoparticle discovery, highlighting the critical role of the parameters discussed.
MLaGA Nanoparticle Discovery Workflow
Parameter Interrelationship Network
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents and Materials for PLGA Nanoparticle Formulation Research
| Reagent/Material | Function in Formulation | Examples & Key Characteristics |
|---|---|---|
| PLGA Polymers | Biodegradable polymer matrix forming the nanoparticle core. | Resomer series (e.g., RG 503H, RG 752H); defined by Mw, LA/GA ratio (e.g., 50:50, 75:25), and end-group (carboxylate vs. ester) [42]. |
| Surfactants & Stabilizers | Stabilize the oil-water interface during emulsion, controlling particle size and preventing aggregation. | Poloxamer 188 (Pluronic F68), Polyvinyl Alcohol (PVA); function characterized by Hydrophilic-Lipophilic Balance (HLB) value [41] [42]. |
| Organic Solvents | Dissolve the polymer and hydrophobic drug for the organic phase. | Acetone (for nanoprecipitation), Dichloromethane (DCM, for emulsion); solvent polarity is a critical feature [41] [40]. |
| Model Active Compounds | Hydrophobic probe molecules used to study encapsulation and release. | Coumarin-6; allows for fluorescence-based tracking and quantification [42]. |
| Computational Tools | Enable in-silico prediction of polymer-drug compatibility and MLaGA-driven optimization. | Molecular Dynamics (MD) simulations for calculating Flory-Huggins (χ) parameter [42]; Gaussian Process Regression (GP) or Support Vector Regression (SVR) as ML surrogates in MLaGA [7] [40]. |
This application note establishes a clear roadmap for leveraging critical input parameters in the design of PLGA nanoparticles. The quantitative relationships and detailed protocols provided enable researchers to move beyond empirical methods. By integrating this knowledge with the power of Machine Learning accelerated Genetic Algorithms, the path to discovering and optimizing novel, high-performance nanocarriers is significantly shortened, marking a new paradigm in data-driven pharmaceutical development.
Navigating the Complexities: Key Challenges and Optimization Strategies for MLaGA
1. Introduction
In the context of machine learning-accelerated genetic algorithms (MLaGAs) for nanoparticle discovery, a primary bottleneck is the scarcity of high-quality, labeled experimental data. The process of synthesizing and characterizing nanoparticles is resource-intensive, resulting in datasets that are too limited for training robust models. This document details practical strategies and protocols to overcome data scarcity, enabling effective model training within a MLaGA framework for drug development research.
2. Quantitative Overview of Data Resampling Techniques
Data resampling techniques artificially adjust the volume and balance of a training dataset. The following table summarizes the core methods.
Table 1: Core Data Resampling Strategies for Imbalanced Datasets
| Strategy | Method | Key Mechanism | Advantages | Disadvantages | Best-Suited Use Case |
|---|---|---|---|---|---|
| Oversampling [44] [45] | Random Oversampling | Duplicates existing minority class examples. | Simple and fast to implement [45]. | High risk of overfitting; models may become too confident [44] [45]. | Very small datasets needing quick balancing [45]. |
| SMOTE (Synthetic Minority Over-sampling Technique) [45] | Generates synthetic examples by interpolating between existing minority class instances. | Creates varied data, not just copies; helps models generalize better [45]. | Can generate noisy samples if data is highly scattered; not for very few initial examples [45]. | Datasets with a decent number of minority examples needing variety [45]. | |
| ADASYN (Adaptive Synthetic) [45] | Focuses on generating samples for minority class examples that are hardest to learn. | Helps models better understand challenging, hard-to-classify regions. | Can over-complicate simple datasets. | Complex datasets with challenging areas [45]. | |
| Undersampling [44] [45] | Random Undersampling | Randomly removes examples from the majority class. | Simple and fast; good for very large datasets [45]. | Potentially discards useful and important information [44] [45]. | Large datasets with redundant majority class examples [45]. |
| Tomek Links [45] | Removes majority class examples that are closest neighbors to minority class examples. | Cleans overlapping data; creates clearer decision boundaries. | Does not reduce the majority class size significantly on its own. | Datasets where classes overlap and need boundary clarification [45]. | |
| ENN (Edited Nearest Neighbors) [45] | Removes any example whose class differs from the class of most of its nearest neighbors. | Effectively removes noise and outliers from both classes. | Can be too aggressive if the data is already clean. | Cleaning messy data and removing outliers [45]. | |
| Hybrid Sampling [45] | SMOTETomek [45] | Applies SMOTE for oversampling, then uses Tomek Links for cleaning. | Balances the dataset while clarifying class boundaries. | Combines the risks of both constituent methods. | Severely imbalanced and noisy datasets [45]. |
| SMOTEENN [45] | Applies SMOTE, then uses ENN to clean both classes. | Can be more aggressive than SMOTETomek in cleaning data. | May lead to an over-optimistic view of model performance. | Datasets requiring both more examples and extensive cleaning [45]. |
3. Advanced Protocol: Synthetic Data Generation using Generative Adversarial Networks (GANs)
For data scarcity beyond simple class imbalance, GANs can generate entirely new, synthetic data points that mimic the underlying distribution of the original, small dataset. This is particularly powerful for creating hypothetical nanoparticle property data.
Table 2: Research Reagent Solutions for GAN-based Data Generation
| Component / Tool | Function / Description | Implementation Consideration |
|---|---|---|
| Generator Network (G) | A neural network that maps a random noise vector to a synthetic data point. Its goal is to "fool" the Discriminator [46]. | Architecture must be complex enough to learn the data distribution but not so large as to overfit the small dataset. |
| Discriminator Network (D) | A neural network that classifies an input data point as "real" (from the original dataset) or "fake" (from the Generator) [46]. | Should be a robust binary classifier; its performance drives the Generator's improvement. |
| Training Dataset | The limited, original dataset of nanoparticle properties (e.g., size, zeta potential, composition, efficacy). | Data must be cleaned and normalized (e.g., using min-max scaling) before training begins [46]. |
| Adversarial Training Loop | The mini-max game where G and D are trained concurrently until D can no longer reliably distinguish real from fake data [46]. | Training can be unstable; techniques like Wasserstein GAN or gradient penalty are often needed for reliable convergence. |
Protocol 3.1: Implementing a GAN for Synthetic Nanoparticle Data Generation
A. Data Preprocessing
- Data Cleaning: Address missing values, which may constitute a small percentage (e.g., ~0.01%) of the dataset, through imputation or removal [46].
- Normalization: Apply min-max scaling to all numerical features to constrain them to a consistent range (e.g., [0, 1]). This ensures stable GAN training [46].
- Formatting: Structure the data into a matrix where each row is a nanoparticle record and each column is a feature.
B. Model Architecture and Training
- Define Networks: Implement the Generator and Discriminator as fully connected or convolutional neural networks using a deep learning framework (e.g., TensorFlow, PyTorch).
- Train the GAN:
- Sample a batch of real data from the preprocessed dataset.
- Sample a batch of random noise vectors.
- Use the Generator to create a batch of synthetic data from the noise.
- Train the Discriminator on the combined batch of real and synthetic data, updating its weights to correctly classify them.
- Train the Generator to produce data that the current Discriminator will classify as "real".
- Iterate: Repeat this adversarial process until the generated data is qualitatively and quantitatively similar to the real data, and the Discriminator's classification accuracy approaches 50% [46].
C. Validation and Integration
- Generate Synthetic Data: Use the trained Generator to create a large synthetic dataset.
- Validate Quality: Employ domain knowledge and statistical tests (e.g., comparing distributions, t-SNE plots) to ensure the synthetic data is realistic and diverse.
- Augment Training Set: Combine the validated synthetic data with the original dataset to form a larger, augmented dataset for downstream MLaGA model training.
4. Visualization of Workflows
Diagram 1: Integrated MLaGA and Data Augmentation Workflow
Diagram 2: GAN Architecture for Synthetic Data Generation
Managing the Exploration-Exploitation Trade-off for Efficient Resource Allocation
Application Note: The MLaGA Framework for Nanoparticle Discovery
Core Principle and Quantitative Performance
The Machine Learning accelerated Genetic Algorithm (MLaGA) is a metaheuristic optimization framework that integrates a machine learning model as a computationally inexpensive surrogate for direct energy calculations. This hybrid approach is designed to manage the exploration-exploitation trade-off efficiently, dramatically reducing the resource expenditure required to discover stable nanoparticle alloys, such as nanoalloy catalysts for clean energy applications [7].
Table 1: Performance Comparison of Search Algorithms for Nanoparticle Discovery. This table summarizes the quantitative efficiency gains achieved by different algorithmic strategies when searching for stable PtxAu147-x nanoparticle homotops [7].
| Algorithm Type | Description | Approx. Number of Energy Calculations | Key Characteristic |
|---|---|---|---|
| Brute-Force | Exhaustive evaluation of all possible configurations | 1.78 x 10⁴⁴ | Computationally infeasible; serves as a theoretical baseline |
| Traditional GA | Evolutionary operations without ML surrogate | ~16,000 | Robust but still computationally expensive |
| Generational MLaGA | ML model used to screen a full generation of candidates | ~1,200 | Enables parallelization of energy calculations |
| Pool-based MLaGA | A new ML model is trained after each energy calculation | ~310 | Serial workflow; minimizes total calculations |
| Pool-based MLaGA with Uncertainty | Exploits model prediction uncertainty for selection | ~280 | Most efficient in terms of total CPU hours |
Managing Exploration and Exploitation
The MLaGA framework explicitly allocates computational resources to balance exploration (searching new regions of the potential energy surface) and exploitation (refining known promising areas) [7]:
- Exploration Mechanism: The traditional Genetic Algorithm component performs crossover and mutation operations, maintaining population diversity and exploring a wide configuration space [7].
- Exploitation Mechanism: The on-the-fly trained Machine Learning surrogate model (e.g., Gaussian Process regression) acts as a cheap energy predictor. It allows for rapid screening and identification of the most promising candidates, focusing expensive computational resources on likely winners [7].
Experimental Protocols
Protocol 1: Traditional Genetic Algorithm for Nanoparticle Optimization
This protocol establishes a baseline for nanoparticle structure search without machine learning acceleration [7].
2.1.1 Research Reagent Solutions
Table 2: Essential Materials and Computational Tools for GA and MLaGA Protocols.
| Item Name | Function / Description | Example / Note |
|---|---|---|
| Template Structure | Defines the initial nanoparticle geometry for the search. | Mackay icosahedral (147-atom) structure [7]. |
| Energy Calculator | Provides the accurate energy evaluation for candidate structures. | Density Functional Theory (DFT) or Effective-Medium Theory (EMT) [7]. |
| Population of Candidates | A set of nanoparticle configurations undergoing evolution. | Typically 150-200 homotops (atomic permutations on a template) [7]. |
| Gaussian Process (GP) Regression Model | The ML surrogate that predicts energies without expensive calculation. | Can be replaced with other ML frameworks (e.g., deep learning) [7]. |
2.1.2 Step-by-Step Methodology
- Initialization: Generate an initial population of candidate nanoparticle structures (homotops) by randomly assigning atomic species (e.g., Pt and Au) to the sites of a predefined template structure (e.g., a 147-atom Mackay icosahedron) [7].
- Evaluation: Calculate the energy (fitness) of every candidate in the population using the chosen energy calculator (e.g., DFT). The energy is typically expressed as an "excess energy" for stability comparison [7].
- Selection: Select parent candidates from the population for reproduction, with a probability weighted by their fitness (lower energy is better) [7].
- Crossover & Mutation: Create a new generation of offspring by:
- Crossover: Swapping segments of atomic coordinates between two parent structures.
- Mutation: Randomly changing the atomic species at a subset of sites in a structure [7].
- Replacement: Form a new population from the best candidates of the parent and offspring pools.
- Convergence Check: Repeat steps 2-5 until the population's fitness shows no significant improvement over multiple generations. For a 147-atom binary nanoalloy, this requires approximately 16,000 energy evaluations to map the convex hull of stable minima [7].
Protocol 2: Machine Learning Accelerated Genetic Algorithm (MLaGA)
This protocol integrates a machine learning surrogate to drastically reduce the number of expensive energy calculations [7].
2.2.1 Step-by-Step Methodology
- Initialization and Initial Evaluation: Generate an initial small population and compute the actual energy for each candidate using DFT to create a seed dataset [7].
- ML Model Training: Train a Gaussian Process (GP) regression model on all candidates for which actual energy has been computed. This model learns the relationship between nanoparticle configuration and its energy [7].
- Nested Surrogate GA (Exploitation):
- Launch a separate, "nested" genetic algorithm that uses the trained GP model as its fitness function.
- This nested GA can run for many generations at a negligible computational cost, as it only requires predictions from the ML model.
- The output is a set of candidate structures predicted by the surrogate to be highly fit [7].
- Tournament Acceptance (Resource Control): From the candidates proposed by the nested GA, select only the top-performing ones (based on ML-predicted fitness) to proceed to actual energy evaluation. This step critically controls resource allocation [7].
- Validation and Model Update: Perform an actual energy calculation (e.g., DFT) on the selected candidates. Add these new data points to the training set and update the ML model [7].
- Convergence Check: The algorithm is considered converged when the ML surrogate cannot propose new candidates predicted to be better than the existing best. This typically occurs after only ~280-1200 DFT calculations, depending on the MLaGA variant used [7].
Diagram 1: Logical workflow of the Machine Learning Accelerated Genetic Algorithm (MLaGA), highlighting the interaction between the master algorithm and the nested surrogate for efficient resource allocation.
Visualization and Implementation Toolkit
Visualizing the Exploration-Exploitation Trade-off
The following diagram models the core decision logic that the MLaGA employs to manage computational resources, dynamically balancing exploration of new regions with exploitation of known promising areas.
Diagram 2: Decision logic for resource allocation in MLaGA, showing how prediction uncertainty and predicted fitness guide the expensive DFT calculations.
Implementing Accessible Visualizations
When generating diagrams for publications or presentations, adherence to accessibility standards is critical. The following guidelines ensure clarity and compliance [47] [48] [49]:
- Color Contrast: For normal text and graphical elements like arrows, ensure a contrast ratio of at least 7:1 against the background. For large-scale text or shapes, a minimum ratio of 4.5:1 is required [47] [48].
- Text within Nodes: The color of text (
fontcolor) inside any node must be explicitly set to have high contrast against the node's fill color (fillcolor) [47]. - Automated Tools: CSS functions like
contrast-color()can automatically select white or black text based on a background color, though manual verification against WCAG guidelines is recommended for mid-tone backgrounds [49].
The development of advanced nanoparticles for drug delivery inherently involves balancing multiple, often competing, objectives. Key properties such as nanoparticle size, drug loading capacity, and therapeutic efficacy frequently exhibit antagonistic relationships; optimizing one typically comes at the expense of another [50] [51]. For instance, while smaller nanoparticles may demonstrate superior tumor penetration and circulation times, they often possess limited volume for drug encapsulation compared to their larger counterparts [51]. Similarly, formulations optimized for maximum drug loading may exhibit reduced release rates, potentially compromising therapeutic bioavailability [50]. These fundamental trade-offs make multi-objective optimization (MOO) an indispensable framework for rational nanoparticle design.
Machine Learning-accelerated Genetic Algorithms (MLaGA) represent a transformative approach to navigating this complex design space. By integrating the robust search capabilities of genetic algorithms with the predictive power of machine learning models, MLaGA enables the rapid identification of Pareto-optimal solutions—formulations where no single objective can be improved without degrading another [7] [9]. This protocol details the application of the MLaGA framework to balance critical antagonistic goals in nanoparticle development, providing researchers with a structured methodology to accelerate the discovery of optimally balanced nanomedicines.
Theoretical Foundation: MLaGA for Nanoparticle Optimization
Core Principles of Machine Learning-Accelerated Genetic Algorithms
The MLaGA framework operates through a synergistic combination of two computational paradigms. Genetic Algorithms (GAs) are population-based optimization methods inspired by Darwinian evolution. They maintain a population of candidate solutions (e.g., nanoparticle formulations) that undergo iterative cycles of evaluation, selection, and variation (through crossover and mutation) to progressively evolve toward better solutions [7]. The Machine Learning component acts as a computationally inexpensive surrogate model, trained on-the-fly to predict the performance of candidate formulations, thereby drastically reducing the number of expensive experimental or computational evaluations required [7] [9].
In practice, the MLaGA implementation often features a two-tiered system [7]:
- A surrogate ML model predicts the fitness of candidates, enabling rapid screening.
- A master GA uses these predictions to guide the evolutionary search, with periodic validation using high-fidelity evaluations (e.g., experimental testing or detailed simulations).
This approach has demonstrated remarkable efficiency gains; for instance, in materials discovery, MLaGA achieved a 50-fold reduction in the number of required energy calculations compared to traditional "brute force" methods [7] [9].
Formulating the Multi-Objective Optimization Problem
For nanoparticle design, the MOO problem can be mathematically formulated as follows [52]: [ \begin{align} \text{Minimize: } & F(x) = [f_1(x), f_2(x), ..., f_k(x)] \ \text{Subject to: } & g_j(x) \leq 0, \quad j = 1, 2, ..., m \ & h_p(x) = 0, \quad p = 1, 2, ..., q \ \end{align} ] where (x) represents a nanoparticle formulation defined by its design variables (e.g., composition, size, surface properties). The functions (f1, f2, ..., fk) represent the conflicting objectives to be minimized (e.g., minimizing size, maximizing drug loading transformed into a minimization problem). The constraints (gj(x)) and (h_p(x)) ensure formulations adhere to critical feasibility criteria, such as synthesis limitations or safety thresholds [52] [50]. The solution to this problem is not a single formulation but a set of Pareto-optimal solutions, collectively known as the Pareto front, which explicitly maps the trade-offs between all objectives.
Figure 1: MLaGA Optimization Workflow. This diagram outlines the iterative process of using a surrogate ML model within a genetic algorithm to efficiently discover Pareto-optimal nanoparticle formulations.
Application Notes: MLaGA for Nanoparticle Development
Defining Objectives and Constraints
The first critical step is to define the key performance objectives and constraints specific to the therapeutic application.
Common Objectives:
- Minimize Nanoparticle Size: Critical for tumor penetration and circulation half-life [51]. Smaller nanoparticles (e.g., ~100-200 nm) often show better extravasation and distribution.
- Maximize Drug Loading/Encapsulation Efficiency: Ensures sufficient therapeutic payload per particle, reducing the required dose and potential side effects [50]. High encapsulation efficiency (e.g., >70%) is often targeted.
- Maximize Therapeutic Efficacy: This can be a direct measure of cell killing in vitro or tumor size reduction in vivo [51]. It is often in conflict with high loading, as dense polymer matrices can hinder drug release.
Typical Constraints:
- Synthesis Feasibility: Limits on polymer concentrations (e.g., PLA: 100-300 mg) or component ratios (e.g., N/P ratio in LNPs) [50] [53].
- Drug-like Criteria: Structural constraints to avoid reactive groups or ensure stability [52].
- Toxicity Thresholds: Maximum allowable excipient concentrations or drug release rates in non-target tissues.
MLaGA in Action: Case Studies
Table 1: Summary of MLaGA Applications in Nanomedicine Optimization
| Case Study | Conflicting Objectives | MLaGA Approach | Key Outcome | Source |
|---|---|---|---|---|
| Polymeric Microcapsules | Maximize Encapsulation Efficiency (Y₁) vs. Maximize Drug Release at 12h (Y₂) | NSGA-III algorithm applied to RSM models linking formulation factors to outcomes. | Identified Pareto front of 5 optimal formulations, revealing inherent trade-off: higher efficiency reduces release rate. | [50] |
| Vasculature-Targeting NPs | Maximize Tumor Accumulation (TNP) vs. Minimize Tumor Diameter (TD) | GA optimized nanoparticle diameter (d) and avidity (α) for a cohort of different-sized tumors. |
Found optimal d for each tumor size; smaller NPs (e.g., 288-334 nm) were superior for larger tumors. |
[51] |
| LNP Formulation (COMET) | Maximize Transfection Efficacy across multiple cell lines | Transformer-based neural network (COMET) used to predict LNP performance from composition and synthesis parameters. | Model accurately ranked LNP efficacy, enabling in-silico screening of 50 million formulations to identify top candidates. | [53] |
Experimental Protocol: Implementing MLaGA for Nanoparticle Optimization
This protocol provides a step-by-step guide for applying the MLaGA framework to optimize a nanoparticle formulation, using a lipid nanoparticle (LNP) system for RNA delivery as a primary example.
Step 1: Problem Formulation and Dataset Generation
- Define Design Variables (x): Identify the controllable parameters of your formulation. For LNPs, this typically includes:
- Ionizable Lipid identity (e.g., C12-200, SM102) and molar percentage.
- Helper Lipid identity (e.g., DOPE, DSPC) and molar percentage.
- Cholesterol and PEG-lipid molar percentages.
- Synthesis parameters (e.g., N/P ratio, aqueous-to-organic phase mixing ratio) [53].
- Define Objectives (F(x)): Specify the properties to be optimized. For RNA-LNPs, the primary objective is often to maximize transfection efficacy (measured by luciferase expression), while potentially minimizing a secondary objective like size or cost.
- Generate Initial Dataset: Create a diverse set of formulations (e.g., 100-500) covering the defined design space. For LNPs, this can be achieved via a high-throughput robotic synthesis platform [53].
- High-Fidelity Evaluation: Experimentally measure all objectives for each formulation in the initial dataset. For efficacy, this involves in vitro transfection assays in relevant cell lines (e.g., DC2.4, B16-F10) [53]. Log-transform and normalize the resulting efficacy data.
Step 2: Building the Machine Learning Surrogate
- Feature Representation: Encode each formulation for the ML model.
- Molecular Structures: Encode lipid SMILES strings into molecular embeddings using a pre-trained chemical language model [53].
- Compositional Features: Create composition embeddings from the molar percentages of each component.
- Synthesis Parameters: Embed continuous parameters like N/P ratio.
- Model Architecture: Implement a model like COMET, which uses a transformer architecture to process the concatenated feature vectors of all components and a formulation-level [CLS] token to make the final prediction [53].
- Model Training:
- Objective: Use a pairwise ranking objective. Instead of predicting absolute efficacy values, the model learns to rank any two LNPs correctly based on their efficacy, which is more robust to experimental noise [53].
- Training Loop: Split the initial dataset (e.g., 70% train, 10% validation, 20% test). Train the model to minimize the ranking loss. Employ ensembling (e.g., 5 models) for improved robustness [53].
Step 3: Genetic Algorithm Setup and Execution
- Population Initialization: Generate an initial population of candidate formulations, either randomly or by sampling from the initial dataset.
- Fitness Evaluation: Use the trained surrogate ML model to predict the fitness (e.g., transfection efficacy rank) of all candidates in the population. This is computationally cheap and allows for rapid screening.
- Selection, Crossover, and Mutation:
- Selection: Use a tournament selection method to choose parent formulations, favoring those with higher predicted fitness [7].
- Crossover: Combine components from two parent formulations to create offspring (e.g., swapping the ionizable lipid type and adjusting molar percentages proportionally).
- Mutation: Randomly modify an offspring's features with a low probability (e.g., change a lipid identity, perturb a molar ratio by ±5%).
- High-Fidelity Validation and Model Update:
- Select the top N predicted candidates from the GA offspring and submit them for experimental validation (Step 1.4).
- Add the new experimental data to the training dataset.
- Periodically retrain the surrogate ML model with the expanded dataset to improve its predictive accuracy. This iterative feedback loop is core to MLaGA's efficiency [7].
Step 4: Pareto Front Analysis and Decision Making
- Identify Non-Dominated Solutions: After the MLaGA run converges, analyze the final population of experimentally validated formulations to identify the Pareto front. A formulation is "non-dominated" if no other formulation is better in all objectives.
- Visualize Trade-offs: Plot the Pareto front, for example, with Encapsulation Efficiency on one axis and Drug Release on the other, to visually illustrate the trade-offs [50].
- Select Final Formulation: The choice from the Pareto-optimal set depends on clinical priorities. A formulation with higher release might be chosen for acute therapy, while one with higher encapsulation might be preferred for sustained release.
Figure 2: Multi-Objective Optimization Logic. This diagram illustrates the fundamental challenge of conflicting objectives and how the MLaGA process identifies a set of optimal compromise solutions (the Pareto front) from which a final formulation is selected.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for MLaGA-Driven Nanoparticle Optimization
| Reagent/Material | Function in Optimization | Example Usage & Rationale |
|---|---|---|
| Biodegradable Polymers (e.g., PLA, PLGA) | Core structural component of polymeric nanoparticles. Concentration and molecular weight are key design variables. | PLA (100-300 mg) forms a compact polymer network. Higher concentrations increase encapsulation efficiency but can hinder drug release [50]. |
| Ionizable Lipids (e.g., C12-200, SM102) | Key functional lipid in LNPs for encapsulating nucleic acids and enabling endosomal escape. Identity and molar % are critical variables. | Different lipids (e.g., C12-200 vs. DLin-MC3-DMA) confer vastly different efficacy. MLaGA can optimize the choice and ratio [53]. |
| Helper Lipids (e.g., DOPE, DSPC) | Modulate the structure and fluidity of the LNP bilayer, influencing stability and fusion with cell membranes. | DOPE tends to promote fusogenicity and enhance efficacy in many cell types, making it a frequent variable [53]. |
| Polyvinylpyrrolidone (PVP K30) | Hydrophilic pore-forming agent in polymeric microspheres. | Increases from 0 to 100 mg accelerate drug release by facilitating dissolution medium penetration, a key variable for release rate optimization [50]. |
| Cholesterol | Stabilizes the LNP bilayer and modulates membrane rigidity and pharmacokinetics. | A nearly universal component, but its molar percentage (typically ~40%) can be optimized by MLaGA for specific applications [53]. |
| PEG-Lipids (e.g., C14-PEG, DMG-PEG) | Shields the LNP surface, reducing opsonization and extending circulation half-life. Impacts size and efficacy. | Molar percentage is a crucial variable; higher PEG content increases size and can reduce efficacy by inhibiting cellular uptake [53]. |
| Firefly Luciferase (FLuc) mRNA | Reporter gene for quantitatively assessing transfection efficacy in high-throughput screening. | Encapsulated in LNPs; bioluminescence readout provides a robust, quantitative measure of functional delivery for training the ML model [53]. |
Application Note
Background and Significance
The discovery and optimization of nanoparticles (NPs) for drug delivery represent a formidable challenge in modern therapeutics, characterized by a vast and complex design space encompassing numerous synthesis parameters and intricate nano-bio interactions [35]. The fusion of molecular-scale engineering in nanotechnology with machine learning (MEA) analytics is reshaping the field of precision medicine [54]. Traditional "brute-force" experimental approaches are often time-consuming, resource-intensive, and lack predictability. The Machine Learning accelerated Genetic Algorithm (MLaGA) paradigm addresses these challenges by merging the robust, global search capabilities of genetic algorithms with the predictive power of machine learning, creating a computationally efficient framework for navigating high-dimensional optimization problems [7]. This protocol details the application of MLaGA for the discovery and formulation of NPs, specifically focusing on achieving predictable and scalable outcomes from initial in-silico simulations to experimentally validated, clinically viable formulations. A key benchmark demonstrates that this approach can yield a 50-fold reduction in the number of required energy calculations compared to a traditional GA, making the exploration of vast compositional and structural spaces, such as binary nanoalloys, feasible using high-fidelity methods like Density Functional Theory (DFT) [9] [16] [7].
The MLaGA framework for nanoparticle discovery integrates a master genetic algorithm with a machine learning surrogate model in an iterative loop. The process begins with the initialization of a population of candidate nanoparticles. A subset of this population is selected for energy evaluation using the primary, computationally expensive calculator. These evaluated candidates are used to train and iteratively improve a machine learning model, which learns to predict the fitness of unevaluated structures. This surrogate model is then deployed within a nested GA to inexpensively screen a vast number of candidate solutions, identifying the most promising individuals. These top candidates are promoted to the master population and evaluated with the primary calculator, closing the loop and providing new data to refine the ML model further. This cycle continues until convergence, efficiently steering the search towards optimal global solutions [7].
Workflow Diagram Title: MLaGA Framework for Nanoparticle Discovery
Protocol: MLaGA for Nanoparticle Formulation Optimization
This protocol provides a step-by-step methodology for implementing the MLaGA to identify stable, compositionally variant nanoparticle alloys, as demonstrated for PtxAu147-x icosahedral particles [7].
Materials and Data Preparation
Research Reagent Solutions
Table 1: Essential Research Reagents and Computational Tools
| Item/Category | Function/Description | Example/Specification |
|---|---|---|
| Genetic Algorithm (GA) Platform | Core optimization engine for evolving candidate solutions. | Custom code or libraries (e.g., in Python) implementing selection, crossover, mutation. |
| Machine Learning (ML) Model | Surrogate for expensive energy calculations; predicts candidate fitness. | Gaussian Process (GP) Regression [7] or Neural Networks [35]. |
| Primary Energy Calculator | High-fidelity method to evaluate the stability/fitness of selected candidates. | Density Functional Theory (DFT) or Effective-Medium Theory (EMT) [7]. |
| Template Structure | Defines the initial geometric configuration for the nanoparticle. | 147-atom Mackay icosahedral structure [7]. |
| Feature Descriptor | Encodes the atomic configuration of a nanoparticle for the ML model. | Composition and local atomic environments [7]. |
Initial Dataset Generation
- Define the Search Space: Specify the compositional range for the binary nanoalloy (e.g., PtxAu147−x for (x \in \left[ {1,146} \right])) [7].
- Initialize Population: Generate an initial population of candidate nanoparticles. This can be a set of random homotops (atomic permutations within a fixed geometric template) for various compositions.
- Calculate Initial Fitness: Select a small, diverse subset of the initial population (e.g., 50-100 candidates) and evaluate their fitness (e.g., excess energy) using the primary energy calculator (e.g., EMT or DFT). This creates the initial training dataset for the ML model.
Computational Procedure
MLaGA Execution
- Master GA Selection: The master GA selects a pool of parent candidates from the current population based on their fitness (actual or predicted).
- Primary Energy Evaluation: Evaluate the selected parents using the primary energy calculator (e.g., DFT) to obtain their accurate fitness. This step is computationally expensive and is therefore performed on a limited number of candidates.
- ML Model Training: Train the surrogate ML model (e.g., a Gaussian Process regressor) on all data collected from primary energy evaluations so far. The model learns to map the feature descriptors of a nanoparticle to its calculated fitness.
- Nested GA Surrogate Search:
- A nested GA is initialized using the current ML model as its fitness evaluator.
- This nested GA performs a full evolutionary search (selection, crossover, mutation) on the surrogate model. Because evaluating the ML model is computationally cheap, this search can explore thousands to millions of candidates very rapidly.
- The goal is to identify candidates that the ML model predicts to have high fitness (low energy).
- Candidate Promotion: The best-performing candidates from the nested GA search are promoted to the master GA population.
- Convergence Check: The cycle repeats from step 1. Convergence is typically achieved when the ML surrogate can no longer find new candidates predicted to be significantly better than the existing ones in the master population, indicating that the global minimum is likely found [7].
Performance Optimization
- Tournament Acceptance: Implement tournament selection in the nested GA to restrict the number of candidates promoted to the master GA. This can significantly reduce the total number of energy calculations required [7].
- Uncertainty Sampling: For pool-based MLaGA, use the ML model's prediction uncertainty (e.g., from Gaussian Processes) to guide the search towards unexplored regions of the design space, improving efficiency [7].
Performance Benchmarking and Validation
The efficiency of the MLaGA must be quantified against traditional methods. Key performance metrics are summarized in the table below.
Table 2: Benchmarking MLaGA Performance for a 147-Atom Nanoalloy Search [7]
| Search Method | Population Type | Key Feature | Approx. Number of Energy Calculations to Find Convex Hull |
|---|---|---|---|
| Traditional GA | Generational | Baseline, no ML | ~16,000 |
| MLaGA | Generational | Nested GA on surrogate model | ~1,200 |
| MLaGA with Tournament Acceptance | Generational | Restricted candidate promotion | < 600 |
| MLaGA (Pool-based) | Pool | Model trained after each calculation | ~310 |
| MLaGA with Uncertainty (Pool-based) | Pool | Utilizes model prediction uncertainty | ~280 |
- DFT Verification: Validate the final predicted stable structures and the convex hull by performing DFT calculations on the top-ranked candidates identified by the MLaGA. This confirms that the search, accelerated by less expensive calculators (e.g., EMT), successfully locates true minima on the high-fidelity potential energy surface [7].
- Hull Analysis: Construct the convex hull of stability by connecting the lowest excess energy of each stable composition. Compositions lying above this hull are deemed unstable [7].
Experimental Validation and Scalability to Clinical Formulations
From Nanoalloys to Drug Delivery Systems
The principles established in optimizing nanoalloys can be translated to the design of drug-loaded nanoparticles. The "fitness" function evolves from thermodynamic stability to encompass critical pharmaceutical properties, such as drug loading efficiency, stability in physiological fluids, targeting specificity, and controlled release profiles [35].
Characterization Workflow for Clinically Viable Formulations
A robust experimental pipeline is essential to validate in-silico predictions and ensure scalability. The following workflow and table outline the key stages and techniques.
Workflow Diagram Title: Experimental Validation Pipeline
Table 3: Key Characterization Techniques for Nanoparticle Formulations
| Characterization Stage | Technique | Key Parameters Measured | Relevance to Clinical Viability |
|---|---|---|---|
| Synthesis & Formulation | Green Synthesis [55] | Monodispersity, shape, size (5-35 nm). | Reduces toxicity, improves biocompatibility. |
| Physicochemical Characterization | UV-Vis Spectroscopy, TEM [55] | NP size, morphology, and dispersion. | Confirms critical quality attributes (CQAs). |
| Nano-Bio Interactions | Laser-Induced Breakdown Spectroscopy (LIBS) [56] | Elemental composition of individual NPs. | Ultra-sensitive quality control. |
| Nano-Bio Interactions | Protein Corona Adsorption Assays [35] | Protein NP interaction, fate in blood. | Predicts stability, biodistribution, and immune response. |
| In Vitro Bio-Evaluation | Cellular Uptake & Cytotoxicity Assays [35] | Internalization efficiency and safety. | Indicates therapeutic potential and initial safety. |
| In Vivo Performance | Biodistribution & Therapeutic Efficacy Studies | Organ accumulation, target engagement, treatment effect. | Ultimate proof-of-concept for efficacy and safety. |
Ensuring Predictability and Scalability
- Data Integrity: The accuracy of the ML model is contingent on the quality and relevance of the training data. Using high-fidelity experimental data for key validation points is crucial for maintaining predictability across the development pipeline [57] [35].
- Explainable AI (XAI): Employing XAI tools like SHAP or LIME can interpret ML model decisions, building trust and providing insights into which NP parameters most influence performance, thereby guiding smarter design [54].
- Manufacturing Considerations: Scalability requires early attention to synthesis parameters. ML can optimize these parameters for reproducibility and cost-effectiveness, while green synthesis methods can enhance safety and environmental sustainability [55].
Benchmarking Success: Validating MLaGA Performance Against State-of-the-Art Methods
The integration of machine learning with genetic algorithms (MLaGA) represents a paradigm shift in computational materials discovery, particularly for the design of nanoparticles for drug delivery. This paradigm combines the robust search capabilities of genetic algorithms (GAs) with the predictive power of machine learning (ML) to accelerate the identification of optimal nanomaterial configurations. Evaluating the performance of such integrated systems requires carefully designed metrics that quantify success across both computational and experimental domains. This article establishes a comprehensive framework of metrics and detailed protocols for researchers developing MLaGA-accelerated nanoparticle systems, with a specific focus on drug delivery applications.
Quantitative Performance Metrics
The performance of MLaGA-driven nanoparticle discovery must be evaluated through a multi-faceted lens that captures computational efficiency, predictive accuracy, and experimental validation. The tables below summarize essential metrics across these domains.
Table 1: Computational Performance Metrics for MLaGA in Nanoparticle Discovery
| Metric Category | Specific Metric | Definition | Interpretation in MLaGA Context |
|---|---|---|---|
| Computational Efficiency | Reduction in Energy Calculations | Ratio of calculations needed vs. traditional GA | MLaGA achieved 50-fold reduction in Pt-Au nanoparticle searches [7] |
| Convergence Profile | Number of evaluations vs. solution quality | MLaGA located convex hull with ~300 vs. 16,000 calculations [7] | |
| CPU Hours | Total computational time | Balance between parallelization and total calculations [7] | |
| Search Quality | Putative Global Minimum Location | Ability to find lowest-energy configuration | Critical for identifying stable nanoparticle alloys [7] |
| Full Convex Hull Mapping | Complete exploration of stable compositions | Essential for PtxAu147−x composition space [7] | |
| Search Space Coverage | Percentage of viable solutions evaluated | MLaGA efficiently navigates 1.78×1044 homotops [7] |
Table 2: Experimental Validation Metrics for Drug-Loaded Nanoparticles
| Metric Category | Specific Metric | Optimal Values | Experimental Significance |
|---|---|---|---|
| Drug Delivery Performance | Encapsulation Efficiency (EE) | R² = 0.96 (Random Forest prediction) [58] | Weight of drug encapsulated per initial drug mass [58] |
| Drug Loading (DL) | R² = 0.93 (Random Forest prediction) [58] | Weight of drug per mass of nanomedicine [58] | |
| Physicochemical Properties | Particle Size | Precise control critical [59] | Influences biodistribution and targeting efficiency [58] |
| Zeta Potential | Key stability indicator [59] | Affects nanoparticle stability and interactions [59] | |
| Size Distribution | Uniform size desired [58] | Microfluidics enable narrow distributions [58] |
Table 3: Machine Learning Model Performance Metrics
| Algorithm | Application Context | Performance | Reference |
|---|---|---|---|
| Random Forest | Predicting PLGA NP EE/DL | R²: 0.96 (EE), 0.93 (DL) [58] | [58] |
| Gaussian Process (GP) Regression | Surrogate energy prediction | Uncertainty estimation via cumulative distribution function [7] | [7] |
| XGBoost | DNA classification for therapeutic targets | >96% accuracy [4] | [4] |
| Support Vector Machine (SVM) | DNA classification for therapeutic targets | <90% accuracy [4] | [4] |
| BAG-SVR | PLGA particle size prediction | Superior performance for size prediction [59] | [59] |
Experimental Protocols
Protocol 1: MLaGA for Nanoalloy Catalyst Discovery
This protocol outlines the methodology for applying Machine Learning Accelerated Genetic Algorithms (MLaGA) to discover stable bimetallic nanoparticle alloys, as demonstrated for PtxAu147-x systems [7].
Materials and Computational Requirements
- Energy Calculator: Density Functional Theory (DFT) or Effective-Medium Theory (EMT) for potential energy surface evaluation [7]
- ML Surrogate Model: Gaussian Process (GP) regression or alternative ML framework (e.g., deep learning) for inexpensive energy prediction [7]
- Template Structure: Atomic template (e.g., 147-atom Mackay icosahedral structure) [7]
- Genetic Algorithm Framework: Implementation supporting crossover, mutation, and selection operations [7]
Procedure
Initialization:
- Define the composition space for the binary alloy system (e.g., (x \in \left[ {1,146} \right]) for PtxAu147−x)
- Generate initial population of candidate homotops (atomic arrangements)
- Calculate initial energies using DFT/EMT calculator
ML Surrogate Training:
- Train GP regression model on initial energy calculations
- Model uses structural descriptors to predict nanoparticle energies
Nested GA Search:
- Implement two-tier evaluation system with ML-predicted fitness and actual energy calculator
- Run nested GA on surrogate model to identify promising candidates
- Use tournament selection to limit candidates passed to master GA
Iterative Refinement:
- Update ML model with new energy calculations
- Balance exploration and exploitation using prediction uncertainty
- Continue until convergence (ML cannot find better candidates)
Validation:
- Verify putative global minima with full DFT calculations
- Map complete convex hull of stable minima
Key Parameters
- Population Size: Generational (e.g., 150) or pool-based [7]
- Convergence Criterion: ML routine unable to find improved candidates [7]
- Performance Target: ~300 energy calculations for full convex hull vs. 16,000 for traditional GA [7]
Protocol 2: Microfluidic Synthesis and Validation of PLGA Nanoparticles
This protocol describes the experimental synthesis and characterization of drug-loaded PLGA nanoparticles optimized through machine learning predictions [58] [59].
Materials
- Polymers: PLGA with varying molecular weights and lactide-to-glycolide (LA/GA) ratios [58]
- Drug Compounds: Therapeutic agents appropriate for encapsulation (e.g., indomethacin) [58]
- Solvents: Organic solvents for polymer dissolution (type and concentration critical) [59]
- Surfactants: Polyvinyl alcohol (PVA) for stabilization [58]
- Microfluidic Device: Chip with specific channel geometry and diameter [58]
Synthesis Procedure
Solution Preparation:
- Dissolve PLGA in appropriate organic solvent at specified concentration (mg/mL)
- Prepare drug solution at desired concentration
- Prepare aqueous surfactant solution (e.g., PVA at specified concentration)
Microfluidic Setup:
- Set flow rates for organic and aqueous phases according to optimized parameters
- Prime microfluidic channels with appropriate solvents
- Establish stable flow conditions before nanoparticle collection
Nanoparticle Formation:
- Introduce organic and aqueous phases into microfluidic device
- Maintain precise control over flow rate ratios and total flow rate
- Collect nanoparticle suspension from outlet channel
Purification:
- Remove organic solvents and free drug via dialysis or centrifugation
- Wash nanoparticles to remove excess surfactant
- Concentrate to desired final concentration
Characterization and Analysis
Size and Distribution:
- Measure hydrodynamic diameter by dynamic light scattering (DLS)
- Analyze polydispersity index (PDI) for size distribution uniformity
Surface Charge:
- Determine zeta potential via electrophoretic light scattering
- Use values to predict colloidal stability
Drug Loading Analysis:
- Quantify encapsulated drug using appropriate analytical method (HPLC, UV-Vis)
- Calculate drug loading (DL) and encapsulation efficiency (EE) using standard formulas [58]
Morphological Examination:
- Analyze nanoparticle morphology using transmission electron microscopy (TEM)
Workflow Visualization
MLaGA Computational-Experimental Workflow
Research Reagent Solutions
Table 4: Essential Materials for MLaGA-Nanoparticle Research
| Category | Specific Item | Function/Role | Example/Notes |
|---|---|---|---|
| Computational Tools | Density Functional Theory (DFT) | Accurate energy calculations for nanoparticle structures [7] | Validates ML predictions; computationally expensive |
| Gaussian Process Regression | ML surrogate for inexpensive energy prediction [7] | Provides uncertainty estimates | |
| Genetic Algorithm Framework | Global optimization of atomic arrangements [7] | Supports crossover, mutation, selection operations | |
| Polymer Materials | PLGA (varied Mw) | Biodegradable nanoparticle matrix [58] [59] | Molecular weight affects drug release profile |
| Polyvinyl Alcohol (PVA) | Stabilizer/surfactant for nanoparticle formation [58] | Concentration critically impacts size | |
| Microfluidic Components | Microfluidic Chips | Controlled nanoparticle synthesis [58] | Channel geometry and diameter affect mixing |
| Flow Control System | Precise manipulation of flow rates [58] | Critical parameter for size control | |
| Characterization Tools | Dynamic Light Scattering | Size and size distribution measurement [59] | Essential quality control |
| Zeta Potential Analyzer | Surface charge and stability assessment [59] | Predicts colloidal stability | |
| HPLC/UV-Vis Spectrometry | Drug loading and encapsulation efficiency quantification [58] | Validates delivery capabilities |
The integration of MLaGA for computational discovery with experimental nanoparticle synthesis represents a powerful paradigm for accelerating drug delivery system development. The metrics and protocols outlined herein provide researchers with a comprehensive framework for quantifying success across both computational and experimental domains. By implementing these standardized evaluation criteria, the field can more effectively compare methodologies, optimize both in silico and experimental processes, and ultimately accelerate the development of advanced nanomedicines. The continued refinement of these metrics will be essential as MLaGA methodologies evolve and find application in increasingly complex nanoparticle systems.
The discovery of new drugs is a time-consuming and expensive process, frequently taking around 15 years with low success rates [60]. Virtual Screening (VS) has emerged as a crucial computational technique to accelerate this process by screening large databases of compounds to identify molecules with similar properties to a given query, thereby reducing the need for extensive experimental characterization [61] [60]. Within VS, Ligand-Based Virtual Screening (LBVS) methods are employed when the structure of the protein target is unknown, relying on the comparison of molecular descriptors such as shape and electrostatic potential [61] [62]. The efficiency of these comparisons is paramount, as molecular databases can contain millions of compounds and are constantly increasing in size [61].
This application note explores the computational efficiency of Tangram CW, a tool conceptualized around the principles of Machine Learning-accelerated Genetic Algorithms (MLaGAs). We frame this within a broader research thesis on MLaGA for nanoparticle discovery, where such algorithms have demonstrated a 50-fold reduction in the number of required energy calculations [7] [9]. We detail the protocol for evaluating Tangram CW's performance against established methods and present quantitative results on its computational efficiency and solution quality. The insights gained are relevant for researchers, scientists, and drug development professionals seeking to optimize their virtual screening workflows.
The MLaGA Principle: From Nanoparticles to Molecular Screening
The core innovation of the Machine Learning-accelerated Genetic Algorithm (MLaGA) is the integration of a machine learning model as a surrogate fitness evaluator within a traditional genetic algorithm framework. This hybrid approach combines the robust global search capabilities of GAs with the rapid predictive power of ML [7].
In computational materials science, this method has been successfully applied to the discovery of stable nanoparticle alloys. A traditional GA requires a large number of expensive energy calculations (e.g., using Density Functional Theory) to evaluate candidate solutions, often around 16,000 evaluations to locate the convex hull of minima for a 147-atom system [7]. The MLaGA overcomes this bottleneck by training an on-the-fly machine learning model (e.g., Gaussian Process regression) on computed data to act as a computationally inexpensive surrogate predictor of energy. This allows for a high-throughput screening of candidates based on their predicted fitness, with only the most promising individuals undergoing full electronic structure calculation [7]. This strategy led to a dramatic reduction in the number of required energy calculations, from ~16,000 to approximately 280-1200, representing a 50-fold increase in efficiency without sacrificing the quality of the solutions found [7] [63].
Tangram CW adapts this MLaGA principle to the problem of ligand-based virtual screening. The "fitness" of a candidate molecule alignment is its similarity score to a query molecule, and the "expensive calculation" is the precise computation of this score, such as the overlapping volume for shape similarity. By using a surrogate model to guide the search, Tangram CW aims to achieve a similar order of magnitude improvement in computational efficiency for navigating vast molecular databases.
Experimental Protocol for Efficiency Benchmarking
Algorithmic Workflow
The following diagram illustrates the core operational workflow of the MLaGA-based Tangram CW system, showcasing the interaction between the genetic algorithm and the machine learning surrogate model.
Detailed Methodology
1. Problem Definition and Objective Function:
- Objective: Maximize the shape or electrostatic similarity between a query molecule and a target molecule from a database.
- Shape Similarity: Quantified as the overlapping volume of their atoms, calculated using a weighted Gaussian algorithm (WEGA) formulation [61] [60]. The Tanimoto Similarity ((T_c)) normalizes this value to a range between 0 (no overlap) and 1 (identical shapes).
- Electrostatic Similarity: Measured by comparing the electrostatic potential of the query and target molecules. This function is notably non-smooth and possesses many local optima, making it a challenging optimization problem [61].
2. Decision Variables and Search Space:
- Each candidate solution in the population represents a specific alignment of the target molecule relative to the query.
- The search space is defined by variables that control this alignment. Tangram CW utilizes a reduced parameter set of six variables: one rotation angle, two three-dimensional coordinates defining the rotation axis (using a semi-sphere parametrization for uniqueness), and three translation values [61]. This is a simplification compared to other algorithms like OptiPharm, which uses ten variables [61].
3. Algorithm Initialization:
- An initial population of candidate alignments is generated, often by centering both molecules at the origin of coordinates [60].
4. Two-Layer Optimization Strategy: Tangram CW implements a two-layer strategy to balance exploration and exploitation [61].
- Layer 1 (Exploration): This stage is designed to broadly explore the search space and identify promising regions that may contain local or global optima. The solutions found here are used to initialize the population for the second layer.
- Layer 2 (Exploitation): This stage focuses on intensifying the search around the promising solutions identified in the first layer, refining the alignments to achieve high accuracy.
5. Guided Search and Convergence Control:
- The evolution of the population is guided by a leader solution (the best-performing individual in each iteration) [61].
- A convergence test is applied to the rotation axis. If convergence is not detected, the algorithm continues to exploit the current axis; otherwise, it explores other axes to avoid stagnation [61].
- Angular variables are handled in a continuous circular way (between (0) and (2\pi)), which more effectively navigates the periodic nature of rotational space.
6. Benchmarking and Validation:
- Reference Algorithms: Tangram CW's performance is benchmarked against state-of-the-art algorithms, notably OptiPharm [61] [60] and WEGA [60].
- Databases: The algorithm is tested on molecular databases of varying sizes (e.g., 1,750 and 28,374 molecules) to assess scalability [61].
- Key Metrics: The primary metrics for comparison are:
- Computational Cost: The total number of similarity function evaluations required per query molecule.
- Solution Quality: The maximum Tanimoto Similarity score ((T_c)) achieved for a given query-target pair.
Results and Performance Analysis
Key Performance Indicators
The implementation of the MLaGA principle and the two-layer guided search in Tangram CW results in significant performance enhancements. The table below summarizes the key quantitative outcomes from its application in virtual screening.
Table 1: Summary of Computational Efficiency and Performance
| Metric | Traditional / Reference Methods | Tangram CW (MLaGA-based) | Improvement / Significance |
|---|---|---|---|
| Function Evaluations | OptiPharm (Baseline) [61] | Up to 87.5 million fewer evaluations per query (on 1,750 molecule DB) and ~6.42 billion fewer (on 28,374 molecule DB) [61] | Drastic reduction in computational effort, enabling faster screening of large databases. |
| Search Space Dimensionality | 10 variables in OptiPharm (rotation angle, axis coordinates, translation) [61] | 6 variables via semi-sphere parametrization [61] | Reduced complexity and avoidance of redundant solutions, enhancing searchability. |
| Solution Quality (Shape) | Comparable to WEGA, which is state-of-the-art in accuracy [60] | Maintains or improves upon the quality of solutions found by OptiPharm [61] | High-fidelity results without compromising accuracy for speed. |
| Solution Quality (Electrostatic) | Standard methods can be easily trapped in local optima [61] | Significant improvements in quality due to design that avoids local optima [61] | Particularly effective for complex, non-smooth objective functions. |
| Underlying Principle Validation | Traditional GA for nanoparticles: ~16,000 energy evaluations [7] | MLaGA for nanoparticles: ~300 energy evaluations [7] | Validates the 50-fold efficiency gain that inspires Tangram CW's design [7] [9]. |
Visualizing the Two-Layer Strategy
The following diagram outlines the two-layer strategy that underpins Tangram CW's efficient search process, balancing global exploration with local refinement.
Table 2: Key Research Reagents and Computational Tools
| Item / Resource | Type | Function in the Virtual Screening Workflow |
|---|---|---|
| Molecular Databases | Data | Large repositories (e.g., ZINC, ChEMBL) containing the 3D structures of millions of compounds to be screened against a query molecule [61]. |
| Query Molecule | Data | The reference compound with known desired properties; the target of the screening is to find molecules that are structurally or electrostatically similar to it [61] [60]. |
| Shape Similarity Function | Software | The objective function that calculates the overlapping volume between molecules, often using a Gaussian model to represent atoms [61] [60]. |
| Electrostatic Similarity Function | Software | The objective function that quantifies the similarity of the electrostatic potential between two molecules, a critical descriptor for biological activity [61]. |
| Genetic Algorithm Core | Software | The optimization engine that manages the population of candidate alignments and applies selection, crossover, and mutation operators to evolve solutions [7] [61]. |
| Machine Learning Surrogate | Software | A trained ML model (e.g., Gaussian Process) that acts as a fast approximation of the expensive similarity function, guiding the GA search [7]. |
| High-Performance Computing (HPC) Cluster | Hardware | Computational infrastructure required to execute the virtual screening algorithm on large databases within a reasonable timeframe [64]. |
This case study demonstrates that the principles of Machine Learning-accelerated Genetic Algorithms, proven highly effective in computational materials discovery [7], can be successfully translated to the domain of virtual screening. Tangram CW, embodying these principles, achieves a dramatic reduction in computational cost—saving billions of evaluations per query—while maintaining or even improving the accuracy of predictions compared to state-of-the-art tools like OptiPharm [61].
The key to this performance lies in its intelligent design: a reduced search space, a two-layer strategy for balanced exploration and exploitation, and the use of a surrogate ML model to avoid costly computations. This efficiency is not achieved at the expense of robustness; in fact, Tangram CW shows a particular aptitude for handling complex objective functions like electrostatic similarity, where it is less prone to becoming trapped in local optima [61].
For researchers in drug development, the implication is clear: the integration of machine learning into evolutionary optimization algorithms presents a viable path toward overcoming the computational bottlenecks associated with screening ever-expanding molecular databases. This acceleration is crucial for shortening drug discovery timelines and bringing new treatments to patients faster. Future work may focus on extending this MLaGA framework to handle flexible molecules and integrating it with multi-objective optimization to simultaneously balance multiple molecular descriptors.
{ dropzone disabled=true }
Comparative Analysis of Optimization Frameworks and Their Applicability to Nanomedicine
The field of nanomedicine faces a significant challenge: the traditional process of designing nanoparticles (NPs) for drug delivery is notoriously inefficient, relying heavily on costly, time-consuming trial-and-error experiments [28] [65]. The high-dimensional design space—encompassing factors such as size, surface chemistry, composition, and payload—makes it difficult to identify optimal formulations that achieve desired pharmacokinetics, biodistribution, and therapeutic efficacy [28] [66]. In recent years, machine learning (ML) and artificial intelligence (AI) have emerged as transformative tools to address these hurdles. This document provides Application Notes and Protocols for applying advanced optimization frameworks, with a specific focus on the Machine Learning Accelerated Genetic Algorithm (MLaGA), to streamline and accelerate nanomedicine discovery for researchers and drug development professionals [28] [7].
Comparative Analysis of Optimization Frameworks
Various computational strategies are being employed to navigate the complex design space of nanomedicines. The table below summarizes the key frameworks, their operating principles, and performance metrics.
Table 1: Comparison of Optimization Frameworks in Nanomedicine
| Framework | Core Principle | Reported Performance/Advantage | Primary Application in Nanomedicine |
|---|---|---|---|
| Traditional Genetic Algorithm (GA) [67] [7] [68] | An evolutionary-inspired metaheuristic that uses selection, crossover, and mutation on a population of candidate solutions. | Required ~16,000 energy evaluations to locate stable nanoparticle configurations in a benchmark study [7]. Robust but computationally expensive. | Structure prediction for atomic/molecular clusters and nanoalloys [7] [68]. |
| Machine Learning Accelerated GA (MLaGA) [7] | A hybrid model where an on-the-fly trained ML model (e.g., Gaussian Process) acts as a surrogate for expensive fitness evaluations. | Achieved a 50-fold reduction in energy calculations (down to ~300-1200) vs. traditional GA [7]. Efficiently explores vast compositional spaces. | Discovery of stable, compositionally variant nanoalloys (e.g., PtxAu147-x) for catalytic applications [7]. |
| Directed Evolution & High-Throughput Screening [28] | Combines physical/virtual compound libraries, DNA barcoding, and ML-driven data analysis in an iterative feedback loop. | Replaces linear screening with iterative, data-driven optimization; rapidly extracts structure-activity relationships [28]. | Rational design of lipid nanoparticles (LNPs) for enhanced mRNA delivery and transfection efficiency [28]. |
| Bat-Optimized ML Models [69] | Uses the Bat Optimization (BA) metaheuristic algorithm to fine-tune hyperparameters of regression models (e.g., KNN). | Optimized KNN model achieved a test R² of 0.944 for predicting PLGA nanoparticle size [69]. Effective for predictive modeling with limited data. | Predicting and optimizing the size and zeta potential of polymeric PLGA nanoparticles for drug delivery [69]. |
| AI-Guided Formulation Platforms [28] | Employs deep learning models like Graph Neural Networks (GNNs) to screen massive chemical libraries in silico. | Platforms like "AGILE" screened 1,200 lipids and extrapolated to 12,000 variants for improved mRNA transfection [28]. | De novo design and screening of novel ionizable lipids for RNA therapeutics [28]. |
Application Notes & Experimental Protocols
Protocol: Implementing an MLaGA for Nanoalloy Discovery
This protocol outlines the methodology for using a Machine Learning Accelerated Genetic Algorithm to discover stable nanoparticle alloys, based on the work of [7].
3.1.1 Research Reagent Solutions & Computational Tools
Table 2: Essential Tools for MLaGA Implementation
| Item | Function/Description |
|---|---|
| Energy Calculator | Software for accurate energy calculation (e.g., Density Functional Theory (DFT) codes). A less accurate method like Effective-Medium Theory (EMT) can be used for initial benchmarking [7]. |
| Machine Learning Library | A library capable of Gaussian Process (GP) regression or other surrogate modeling (e.g., scikit-learn, GPy) [7]. |
| Genetic Algorithm Framework | A flexible codebase for implementing GA operators (crossover, mutation) and population management. Can be custom-built or adapted from existing packages [7] [68]. |
| Template Structures | Initial atomic coordinate files for the nanoparticle structure to be optimized (e.g., Mackay icosahedron for 147-atom particles) [7]. |
3.1.2 Step-by-Step Workflow
- Problem Initialization: Define the nanoparticle system, including the template structure and the range of chemical compositions to be explored (e.g., PtxAu147-x for (x \in \left[ {1,146} \right])) [7].
- Initial Population Generation: Create a first generation of candidate structures by randomly assigning atoms to the template sites according to the desired composition [7] [68].
- Fitness Evaluation (Initial): Calculate the true fitness (e.g., excess energy) of each candidate in the initial population using the high-fidelity energy calculator (DFT) [7].
- ML Model Training: Train a surrogate ML model (e.g., Gaussian Process regressor) on the collected data of candidate structures and their calculated energies [7].
- Nested Surrogate GA: a. Within the main ("master") GA, run a nested GA where the fitness of candidates is evaluated using the trained ML model, not the expensive calculator. b. The final population from this nested search returns promising, unrelaxed candidates to the master GA [7].
- Selection & New Generation: Select the fittest candidates (based on true energy calculations) and use genetic operators (crossover, mutation) to create a new generation. [68] suggests dynamic management of operator rates based on performance.
- Iteration and Convergence: Repeat steps 4-6. Convergence is typically achieved when the ML surrogate can no longer find new candidates predicted to be better than the existing population, indicating a stable solution has been found [7].
The following workflow diagram illustrates this iterative MLaGA process.
Protocol: ML-Driven Optimization of Polymeric Nanoparticles
This protocol details a methodology for using machine learning to optimize the synthesis parameters of polymeric nanoparticles like PLGA, based on the approach of [69].
3.2.1 Research Reagent Solutions & Computational Tools
Table 3: Essential Tools for ML-Driven Nanoparticle Optimization
| Item | Function/Description |
|---|---|
| Experimental Dataset | A curated dataset of synthesis parameters (e.g., polymer type, antisolvent type, concentrations) and resulting NP properties (size, zeta potential) [69]. |
| ML Library with Preprocessing | A library such as scikit-learn containing KNN, ensemble methods (Bagging, AdaBoost), and preprocessing tools for encoding and outlier detection [69]. |
| Optimization Algorithm | An implementation of the Bat Optimization Algorithm (BA) or similar metaheuristic for hyperparameter tuning [69]. |
| Generative Model (Optional) | A Generative Adversarial Network (GAN) framework for synthetic data generation to augment small datasets, as used in the SBNNR model [69]. |
3.2.2 Step-by-Step Workflow
- Data Preprocessing: a. Categorical Encoding: Transform categorical variables (e.g., polymer type, antisolvent type) into numerical values using Leave-One-Out (LOO) encoding [69]. b. Outlier Detection: Identify and handle anomalies in the dataset using the Local Outlier Factor (LOF) algorithm [69].
- Hyperparameter Optimization: Utilize the Bat Optimization Algorithm (BA) to find the optimal hyperparameters for the chosen regression model (e.g., K-Nearest Neighbors) [69].
- Model Training & Validation: Train the final model (e.g., BA-optimized KNN, or the novel SBNNR model) on the preprocessed training data. Validate its performance using metrics like R² on a held-out test set [69].
- Prediction & Optimization: Use the trained model to predict nanoparticle properties (size, zeta potential) for new, untested combinations of synthesis parameters. Iterate to identify the parameter set that produces the desired NP characteristics.
The logical relationship of this data-driven pipeline is shown below.
The integration of advanced computational frameworks, particularly Machine Learning Accelerated Genetic Algorithms (MLaGAs), is ushering in a new paradigm for rational design in nanomedicine. As summarized in this document, these methods offer dramatic improvements in efficiency—reducing the number of required experiments or simulations by orders of magnitude—while effectively navigating the complex, high-dimensional design spaces of nanoparticles [28] [7]. The provided protocols for MLaGA and ML-driven optimization offer researchers actionable methodologies to implement these powerful approaches. By moving beyond traditional trial-and-error, these data-driven strategies hold the potential to significantly accelerate the discovery and development of next-generation nanomedicines, from optimized polymeric carriers to novel nanoalloy catalysts [28] [7] [69].
Conclusion
The integration of Machine Learning with Genetic Algorithms represents a paradigm shift in nanoparticle discovery, directly addressing the critical bottlenecks of time, cost, and complexity in biomedical research. By synthesizing the key intents, it is evident that MLaGA provides a robust foundational framework, practical methodological workflows, effective troubleshooting approaches, and validated performance superior to traditional methods. The future of this field lies in the development of more automated, closed-loop systems that tightly integrate computational prediction with robotic synthesis and high-throughput screening. As ML models become more sophisticated and datasets more expansive, MLaGA is poised to move beyond optimizing known parameters to generating novel nanoparticle designs, ultimately accelerating the translation of nanomedicines from the laboratory to the clinic and enabling truly personalized therapeutic solutions.
References
- 1. The Quest for the Ultimate Algorithm III — The Niche of ... [https://medium.com/@adnanmasood/the-quest-for-the-ultimate-algorithm-iii-the-niche-of-global-search-and-optimization-with-cb5ef8d742ac]
- 2. AI-guided platform improves design and efficiency of ... [https://www.news-medical.net/news/20250924/AI-guided-platform-improves-design-and-efficiency-of-therapeutic-nanoparticles.aspx]
- 3. Optimizing imbalanced learning with genetic algorithm [https://www.nature.com/articles/s41598-025-09424-x]
- 4. Comparison of classical Machine Learning-based ... [https://www.sciencedirect.com/science/article/pii/S2950363924000139]
- 5. Nanoparticles of natural product-derived medicines [https://www.sciencedirect.com/science/article/pii/S240584402501120X]
- 6. Challenges and opportunities in computational studies for ... [https://www.nature.com/articles/s44386-025-00024-3]
- 7. Genetic algorithms for computational materials discovery ... [https://www.nature.com/articles/s41524-019-0181-4]
- 8. A Machine Learning Approach for PLGA Nanoparticles in ... [https://www.mdpi.com/1999-4923/15/2/495]
- 9. Machine Learning Accelerated Genetic Algorithms ... [http://www.tri.global/research/machine-learning-accelerated-genetic-algorithms-computational-materials-search]
- 10. Material informatics-driven insights into brain cancer ... [https://www.sciencedirect.com/science/article/pii/S2352952024000264]
- 11. A formulation dataset of poly(lactide-co-glycolide) ... [https://pubmed.ncbi.nlm.nih.gov/40640221/]
- 12. Current and Near-Future Technologies to Quantify ... [https://www.mdpi.com/2306-5354/12/4/362]
- 13. Cluster-MLP: An Active Learning Genetic Algorithm ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10598790/]
- 14. A Machine Learning Approach for PLGA Nanoparticles in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9966002/]
- 15. Surrogate model [https://en.wikipedia.org/wiki/Surrogate_model]
- 16. Machine Learning Accelerated Genetic Algorithms for ... [https://chemrxiv.org/engage/chemrxiv/article-details/60c73fdfbb8c1a018b3d9c5f]
- 17. Surrogate Based Genetic Algorithm Method for Efficient ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9933449/]
- 18. Surrogate models in evolutionary single-objective ... [https://www.sciencedirect.com/science/article/pii/S0020025521002395]
- 19. Multi-objective optimization [https://en.wikipedia.org/wiki/Multi-objective_optimization]
- 20. Multi-Objective Optimization (MOO) | by Chong Jing Ting [https://medium.com/@chongjingting/multi-objective-optimization-moo-1b29ece5b64f]
- 21. Multiobjective optimization [https://openmdao.github.io/PracticalMDO/Notebooks/Optimization/multiobjective.html]
- 22. Nanoparticle - an overview [https://www.sciencedirect.com/topics/materials-science/nanoparticle]
- 23. Nanoparticle [https://en.wikipedia.org/wiki/Nanoparticle]
- 24. The History of Nanoscience and Nanotechnology [https://pmc.ncbi.nlm.nih.gov/articles/PMC6982820/]
- 25. Machine Learning Accelerated Genetic Algorithms for ... [https://chemrxiv.org/articles/Machine_Learning_Accelerated_Genetic_Algorithms_for_Computational_Materials_Search/7411172/files/13710383.pdf]
- 26. Machine Learning-Driven Innovations in Microfluidics - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11674507/]
- 27. Combining machine learning with high-content imaging to ... [https://www.nature.com/articles/s41467-024-49433-4]
- 28. Machine Learning and Artificial Intelligence in Nanomedicine [https://pmc.ncbi.nlm.nih.gov/articles/PMC12353477/]
- 29. A Novel Active Learning Regression Framework for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7515147/]
- 30. Machine learning-guided high throughput nanoparticle design [https://pubs.rsc.org/en/content/articlehtml/2024/dd/d4dd00104d]
- 31. Balancing the exploration-exploitation trade-off in active ... [https://arxiv.org/abs/2508.18170]
- 32. The Influence of Genetic Algorithms on Learning ... [https://www.mdpi.com/2073-431X/11/5/70]
- 33. PEGylated PLGA nanoparticles: unlocking advanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12288292/]
- 34. Multifunctional PLGA nanosystems: enabling integrated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488692/]
- 35. Machine Learning‐Enhanced Nanoparticle Design for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12376635/]
- 36. Gold nanoparticle-loaded multifunctional Ti3C2Tx MXene ... [https://www.sciencedirect.com/science/article/abs/pii/S1748013225002208]
- 37. Stimuli-Responsive MXene Nanomaterials for Advanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12595973/]
- 38. Bioactive surface-functionalized MXenes for biomedicine [https://pubs.rsc.org/en/content/articlehtml/2025/nr/d4nr04260c]
- 39. Poly(lactic-co-glycolic acid) microsphere production based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8245074/]
- 40. Machine learning assisted exploration of the influential ... [https://www.nature.com/articles/s41598-023-50876-w]
- 41. A formulation dataset of poly(lactide-co-glycolide ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12246394/]
- 42. Influence of lactide vs glycolide composition of poly (lactic- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8484083/]
- 43. Identification and Characterization of Critical Processing ... [https://www.mdpi.com/1999-4923/16/6/796]
- 44. Random Oversampling and Undersampling for Imbalanced ... [https://www.machinelearningmastery.com/random-oversampling-and-undersampling-for-imbalanced-classification/]
- 45. Oversampling and Undersampling, Explained: A Visual ... [https://towardsdatascience.com/oversampling-and-undersampling-explained-a-visual-guide-with-mini-2d-dataset-1155577d3091/]
- 46. Strategies for overcoming data scarcity, imbalance, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11053123/]
- 47. Text has enhanced contrast | ACT Rule | WAI [https://www.w3.org/WAI/standards-guidelines/act/rules/09o5cg/]
- 48. Text has enhanced contrast - Rules - Siteimprove [https://alfa.siteimprove.com/rules/sia-r66]
- 49. contrast-color() - CSS - MDN Web Docs [https://developer.mozilla.org/en-US/docs/Web/CSS/Reference/Values/color_value/contrast-color]
- 50. Trade-offs in Encapsulation Efficiency and Drug Release [https://www.sciepublish.com/article/pii/344]
- 51. Multi-objective optimization of tumor response to drug ... [https://www.nature.com/articles/s41598-020-65162-2]
- 52. CMOMO: a deep multi-objective optimization framework for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12240737/]
- 53. Designing lipid nanoparticles using a transformer-based ... [https://www.nature.com/articles/s41565-025-01975-4]
- 54. Nanotechnology and machine learning: a promising ... [https://www.sciencedirect.com/science/article/pii/S2666521225000717]
- 55. Green Synthesis of Silver Nanoparticles and Its ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10054154/]
- 56. Detection and Identification of Individual Nanoparticles ... [https://www.spectroscopyonline.com/view/detection-and-identification-individual-nanoparticles-using-laser-induced-breakdown-spectroscopy-lib]
- 57. Artificial intelligence in cardiovascular pharmacotherapy [https://pmc.ncbi.nlm.nih.gov/articles/PMC12488325/]
- 58. Intelligence prediction of microfluidically prepared ... [https://www.nature.com/articles/s41598-025-21471-y]
- 59. Design of Poly(lactic-co-glycolic acid) nanoparticles in drug ... [https://www.sciencedirect.com/science/article/abs/pii/S0169743925000206]
- 60. An evolutionary algorithm to compare shape similarity - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6361934/]
- 61. A two-layer mono-objective algorithm based on guided ... [https://www.nature.com/articles/s41598-022-16913-w]
- 62. On the application of artificial intelligence in virtual screening [https://pubmed.ncbi.nlm.nih.gov/40388244/]
- 63. Genetic algorithms for computational materials discovery ... [https://orbit.dtu.dk/en/publications/genetic-algorithms-for-computational-materials-discovery-accelera]
- 64. MultiPharm-DT: A Multi-Objective Decision Tool for Ligand- ... [https://informatica.vu.lt/journal/INFORMATICA/article/1240]
- 65. Machine learning reshapes the paradigm of nanomedicine ... [https://www.sciencedirect.com/science/article/pii/S2211383525003211]
- 66. Rational nanoparticle design: Optimization using insights ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365923004418]
- 67. Genetic Algorithm-Based Design for Metal-Enhanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6600714/]
- 68. A New Genetic Algorithm Approach Applied to Atomic and ... [https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2019.00707/full]
- 69. Utilization of machine learning approach for production ... [https://www.nature.com/articles/s41598-025-92725-y]